AI26 inhibits the ADP-ribosylhydrolase ARH3 and suppresses DNA damage repair
- PMID: 32753484
- PMCID: PMC7535916
- DOI: 10.1074/jbc.RA120.012801
AI26 inhibits the ADP-ribosylhydrolase ARH3 and suppresses DNA damage repair
Abstract
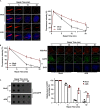
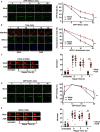
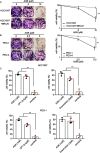
The ADP-ribosylhydrolase ARH3 plays a key role in DNA damage repair, digesting poly(ADP-ribose) and removing ADP-ribose from serine residues of the substrates. Specific inhibitors that selectively target ARH3 would be a useful tool to examine DNA damage repair, as well as a possible strategy for tumor suppression. However, efforts to date have not identified any suitable compounds. Here, we used in silico and biochemistry screening to search for ARH3 inhibitors. We discovered a small molecule compound named ARH3 inhibitor 26 (AI26) as, to our knowledge, the first ARH3 inhibitor. AI26 binds to the catalytic pocket of ARH3 and inhibits the enzymatic activity of ARH3 with an estimated IC50 of ∼2.41 μm in vitro Moreover, hydrolysis of DNA damage-induced ADP-ribosylation was clearly inhibited when cells were pretreated with AI26, leading to defects in DNA damage repair. In addition, tumor cells with DNA damage repair defects were hypersensitive to AI26 treatment, as well as combinations of AI26 and other DNA-damaging agents such as camptothecin and doxorubicin. Collectively, these results reveal not only a chemical probe to study ARH3-mediated DNA damage repair but also a chemotherapeutic strategy for tumor suppression.
Keywords: ADP-ribosylation; ARH3; DNA damage response; DNA repair; cancer therapy; dePARylation; inhibitor.
© 2020 Liu et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

