PPP6C Negatively Regulates STING-Dependent Innate Immune Responses
- PMID: 32753499
- PMCID: PMC7407089
- DOI: 10.1128/mBio.01728-20
PPP6C Negatively Regulates STING-Dependent Innate Immune Responses
Abstract
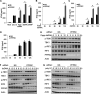
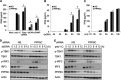
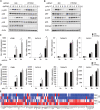
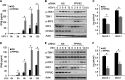
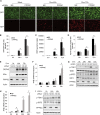
Stimulator of interferon genes (STING) is an essential adaptor protein of the innate DNA-sensing signaling pathway, which recognizes genomic DNA from invading pathogens to establish antiviral responses in host cells. STING activity is tightly regulated by several posttranslational modifications, including phosphorylation. However, specifically how the phosphorylation status of STING is modulated by kinases and phosphatases remains to be fully elucidated. In this study, we identified protein phosphatase 6 catalytic subunit (PPP6C) as a binding partner of Kaposi's sarcoma-associated herpesvirus (KSHV) open reading frame 48 (ORF48), which is a negative regulator of the cyclic GMP-AMP synthase (cGAS)-STING pathway. PPP6C depletion enhances double-stranded DNA (dsDNA)-induced and 5'ppp double-stranded RNA (dsRNA)-induced but not poly(I:C)-induced innate immune responses. PPP6C negatively regulates dsDNA-induced IRF3 activation but not NF-κB activation. Deficiency of PPP6C greatly inhibits the replication of herpes simplex virus 1 (HSV-1) and vesicular stomatitis virus (VSV) as well as the reactivation of KSHV, due to increased type I interferon production. We further demonstrated that PPP6C interacts with STING and that loss of PPP6C enhances STING phosphorylation. These data demonstrate the important role of PPP6C in regulating STING phosphorylation and activation, which provides an additional mechanism by which the host responds to viral infection.IMPORTANCE Cytosolic DNA, which usually comes from invading microbes, is a dangerous signal to the host. The cGAS-STING pathway is the major player that detects cytosolic DNA and then evokes the innate immune response. As an adaptor protein, STING plays a central role in controlling activation of the cGAS-STING pathway. Although transient activation of STING is essential to trigger the host defense during pathogen invasion, chronic STING activation has been shown to be associated with several autoinflammatory diseases. Here, we report that PPP6C negatively regulates the cGAS-STING pathway by removing STING phosphorylation, which is required for its activation. Dephosphorylation of STING by PPP6C helps prevent the sustained production of STING-dependent cytokines, which would otherwise lead to severe autoimmune disorders. This work provides additional mechanisms on the regulation of STING activity and might facilitate the development of novel therapeutics designed to prevent a variety of autoinflammatory disorders.
Keywords: HSV-1; KSHV; Kaposi's sarcoma-associated herpesvirus; PP6C; PPP6C; STING; VSV; herpes simplex virus; interferon; interferons; phosphatase; phosphorylation; protein phosphorylation; vesicular stomatitis virus.
Copyright © 2020 Ni et al.
Figures

References
-
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159:1563–1577. doi: 10.1016/j.cell.2014.11.037. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

