Selective dysregulation of ROCK2 activity promotes aberrant transcriptional networks in ABC diffuse large B-cell lymphoma
- PMID: 32753663
- PMCID: PMC7403583
- DOI: 10.1038/s41598-020-69884-1
Selective dysregulation of ROCK2 activity promotes aberrant transcriptional networks in ABC diffuse large B-cell lymphoma
Abstract
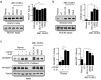
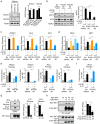
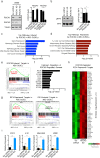
Activated B-cell-like diffuse large B-cell lymphoma (ABC-DLBCL) is an aggressive subtype of lymphoma usually associated with inferior outcomes. ABC-DLBCL exhibits plasmablastic features and is characterized by aberrancies in the molecular networks controlled by IRF4. The signaling pathways that are dysregulated in ABC-DLBCL are, however, not fully understood. ROCK2 is a serine-threonine kinase whose role in lymphomagenesis is unknown. Here we show that ROCK2 activity is constitutively dysregulated in ABC-DLBCL but not in GCB-DLBCL and BL. We furthermore show that ROCK2 phosphorylates IRF4 and that the ROCK2-mediated phosphorylation of IRF4 modulates its ability to regulate a subset of target genes. In addition to its effects on IRF4, ROCK2 also controls the expression of MYC in ABC-DLBCL by regulating MYC protein levels. ROCK inhibition furthermore selectively decreases the proliferation and survival of ABC-DLBCL in vitro and inhibits ABC-DLBCL growth in xenograft models. Thus, dysregulated ROCK2 activity contributes to the aberrant molecular program of ABC-DLBCL via its dual ability to modulate both IRF4- and MYC-controlled gene networks and ROCK inhibition could represent an attractive therapeutic target for the treatment of ABC-DLBCL.
Conflict of interest statement
ABP previously received an investigator-initiated research grant from Kadmon Corporation for studies of SLE T-cells. All other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

