Systematized reporter assays reveal ZIC protein regulatory abilities are Subclass-specific and dependent upon transcription factor binding site context
- PMID: 32753700
- PMCID: PMC7403390
- DOI: 10.1038/s41598-020-69917-9
Systematized reporter assays reveal ZIC protein regulatory abilities are Subclass-specific and dependent upon transcription factor binding site context
Abstract
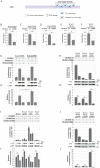
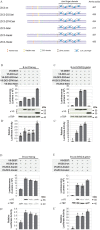
The ZIC proteins are a family of transcription regulators with a well-defined zinc finger DNA-binding domain and there is evidence that they elicit functional DNA binding at a ZIC DNA binding site. Little is known, however, regarding domains within ZIC proteins that confer trans-activation or -repression. To address this question, a new cell-based trans-activation assay system suitable for ZIC proteins in HEK293T cells was constructed. This identified two previously unannotated evolutionarily conserved regions of ZIC3 that are necessary for trans-activation. These domains are found in all Subclass A ZIC proteins, but not in the Subclass B proteins. Additionally, the Subclass B proteins fail to elicit functional binding at a multimerised ZIC DNA binding site. All ZIC proteins, however, exhibit functional binding when the ZIC DNA binding site is embedded in a multiple transcription factor locus derived from ZIC target genes in the mouse genome. This ability is due to several domains, some of which are found in all ZIC proteins, that exhibit context dependent trans-activation or -repression activity. This knowledge is valuable for assessing the likely pathogenicity of variant ZIC proteins associated with human disorders and for determining factors that influence functional transcription factor binding.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Ali RG, Bellchambers HM, Arkell RM. Zinc fingers of the cerebellum (Zic): Transcription factors and co-factors. Int. J. Biochem. Cell Biol. 2012;44:2065–2068. - PubMed
-
- Aruga J, et al. A wide-range phylogenetic analysis of Zic proteins: Implications for correlations between protein structure conservation and body plan complexity. Genomics. 2006;87:783–792. - PubMed
-
- Koyabu Y, Nakata K, Mizugishi K, Aruga J, Mikoshiba K. Physical and Functional Interactions between Zic and Gli Proteins. J. Biol. Chem. 2001;276:6889–6892. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

