Profiling the Course of Resolving vs. Persistent Inflammation in Human Monocytes: The Role of IL-1 Family Molecules
- PMID: 32754155
- PMCID: PMC7365847
- DOI: 10.3389/fimmu.2020.01426
Profiling the Course of Resolving vs. Persistent Inflammation in Human Monocytes: The Role of IL-1 Family Molecules
Abstract
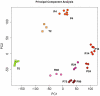
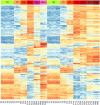
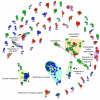
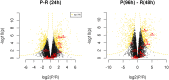
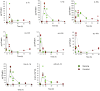
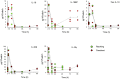
Monocytes and macrophages have a central role in all phases of an inflammatory reaction. To understanding the regulation of monocyte activation during a physiological or pathological inflammation, we propose two in vitro models that recapitulate the different phases of the reaction (recruitment, initiation, development, and resolution vs. persistence of inflammation), based on human primary blood monocytes exposed to sequential modifications of microenvironmental conditions. These models exclusively describe the functional development of blood-derived monocytes that first enter an inflammatory site. All reaction phases were profiled by RNA-Seq, and the two models were validated by studying the modulation of IL-1 family members. Genes were differentially modulated, and distinct clusters were identified during the various phases of inflammation. Pathway analysis revealed that both models were enriched in pathways involved in innate immune activation. We observe that monocytes acquire an M1-like profile during early inflammation, and switch to a deactivated M2-like profile during both the resolving and persistent phases. However, during persistent inflammation they partially maintain an M1 profile, although they lose the ability to produce inflammatory cytokines compared to M1 cells. The production of IL-1 family molecules by ELISA reflected the transcriptomic profiles in the distinct phases of the two inflammatory reactions. Based on the results, we hypothesize that persistence of inflammatory stimuli cannot maintain the M1 activated phenotype of incoming monocytes for long, suggesting that the persistent presence of M1 cells and effects in a chronically inflamed tissue is mainly due to activation of newly incoming cells. Moreover, being IL-1 family molecules mainly expressed and secreted by monocytes during the early stages of the inflammatory response (within 4-14 h), and the rate of their production decreasing during the late phase of both resolving and persistent inflammation, we suppose that IL-1 factors are key regulators of the acute defensive innate inflammatory reaction that precedes establishment of longer-term adaptive immunity, and are mainly related to the presence of recently recruited blood monocytes. The well-described role of IL-1 family cytokines and receptors in chronic inflammation is therefore most likely dependent on the continuous influx of blood monocytes into a chronically inflamed site.
Keywords: IL-1 family; in vitro model; inflammation; macrophages; monocytes.
Copyright © 2020 Italiani, Mosca, Della Camera, Melillo, Migliorini, Milanesi and Boraschi.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

