Interpretable Clinical Genomics with a Likelihood Ratio Paradigm
- PMID: 32755546
- PMCID: PMC7477017
- DOI: 10.1016/j.ajhg.2020.06.021
Interpretable Clinical Genomics with a Likelihood Ratio Paradigm
Abstract
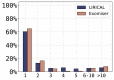
Human Phenotype Ontology (HPO)-based analysis has become standard for genomic diagnostics of rare diseases. Current algorithms use a variety of semantic and statistical approaches to prioritize the typically long lists of genes with candidate pathogenic variants. These algorithms do not provide robust estimates of the strength of the predictions beyond the placement in a ranked list, nor do they provide measures of how much any individual phenotypic observation has contributed to the prioritization result. However, given that the overall success rate of genomic diagnostics is only around 25%-50% or less in many cohorts, a good ranking cannot be taken to imply that the gene or disease at rank one is necessarily a good candidate. Here, we present an approach to genomic diagnostics that exploits the likelihood ratio (LR) framework to provide an estimate of (1) the posttest probability of candidate diagnoses, (2) the LR for each observed HPO phenotype, and (3) the predicted pathogenicity of observed genotypes. LIkelihood Ratio Interpretation of Clinical AbnormaLities (LIRICAL) placed the correct diagnosis within the first three ranks in 92.9% of 384 case reports comprising 262 Mendelian diseases, and the correct diagnosis had a mean posttest probability of 67.3%. Simulations show that LIRICAL is robust to many typically encountered forms of genomic and phenomic noise. In summary, LIRICAL provides accurate, clinically interpretable results for phenotype-driven genomic diagnostics.
Keywords: Human Phenotype Ontology; exome sequencing; genome sequencing; liklihood ratio; phenotype-driven genomic diagnostics.
Copyright © 2020 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
P.N.R. has filed a patent application based on this work.
Figures




References
-
- Sifrim A., Popovic D., Tranchevent L.-C., Ardeshirdavani A., Sakai R., Konings P., Vermeesch J.R., Aerts J., De Moor B., Moreau Y. eXtasy: variant prioritization by genomic data fusion. Nat. Methods. 2013;10:1083–1084. - PubMed
-
- Singleton M.V., Guthery S.L., Voelkerding K.V., Chen K., Kennedy B., Margraf R.L., Durtschi J., Eilbeck K., Reese M.G., Jorde L.B. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet. 2014;94:599–610. - PMC - PubMed
-
- Javed A., Agrawal S., Ng P.C. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat. Methods. 2014;11:935–937. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

