SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness
- PMID: 32756549
- PMCID: PMC7581537
- DOI: 10.1038/s41586-020-2622-0
SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness
Abstract
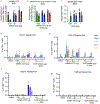
A vaccine for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is needed to control the coronavirus disease 2019 (COVID-19) global pandemic. Structural studies have led to the development of mutations that stabilize Betacoronavirus spike proteins in the prefusion state, improving their expression and increasing immunogenicity1. This principle has been applied to design mRNA-1273, an mRNA vaccine that encodes a SARS-CoV-2 spike protein that is stabilized in the prefusion conformation. Here we show that mRNA-1273 induces potent neutralizing antibody responses to both wild-type (D614) and D614G mutant2 SARS-CoV-2 as well as CD8+ T cell responses, and protects against SARS-CoV-2 infection in the lungs and noses of mice without evidence of immunopathology. mRNA-1273 is currently in a phase III trial to evaluate its efficacy.
Conflict of interest statement
Competing Interest Declaration
K.S.C., N.W., J.S.M., and B.S.G. are inventors on International Patent Application No. WO/2018/081318 entitled “Prefusion Coronavirus Spike Proteins and Their Use.” K.S.C., O.M.A., G.B.H., N.W., D.W., J.S.M, and B.S.G. are inventors on US Patent Application No. 62/972,886 entitled “2019-nCoV Vaccine”. R.S.B. filed an invention report for the SARS-CoV-2 MA virus (UNC ref. #18752).
Figures







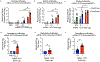

Update of
-
SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness.bioRxiv [Preprint]. 2020 Jun 11:2020.06.11.145920. doi: 10.1101/2020.06.11.145920. bioRxiv. 2020. Update in: Nature. 2020 Oct;586(7830):567-571. doi: 10.1038/s41586-020-2622-0. PMID: 32577634 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

