Genome-wide Modeling of Polygenic Risk Score in Colorectal Cancer Risk
- PMID: 32758450
- PMCID: PMC7477007
- DOI: 10.1016/j.ajhg.2020.07.006
Genome-wide Modeling of Polygenic Risk Score in Colorectal Cancer Risk
Abstract
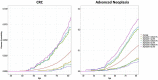
Accurate colorectal cancer (CRC) risk prediction models are critical for identifying individuals at low and high risk of developing CRC, as they can then be offered targeted screening and interventions to address their risks of developing disease (if they are in a high-risk group) and avoid unnecessary screening and interventions (if they are in a low-risk group). As it is likely that thousands of genetic variants contribute to CRC risk, it is clinically important to investigate whether these genetic variants can be used jointly for CRC risk prediction. In this paper, we derived and compared different approaches to generating predictive polygenic risk scores (PRS) from genome-wide association studies (GWASs) including 55,105 CRC-affected case subjects and 65,079 control subjects of European ancestry. We built the PRS in three ways, using (1) 140 previously identified and validated CRC loci; (2) SNP selection based on linkage disequilibrium (LD) clumping followed by machine-learning approaches; and (3) LDpred, a Bayesian approach for genome-wide risk prediction. We tested the PRS in an independent cohort of 101,987 individuals with 1,699 CRC-affected case subjects. The discriminatory accuracy, calculated by the age- and sex-adjusted area under the receiver operating characteristics curve (AUC), was highest for the LDpred-derived PRS (AUC = 0.654) including nearly 1.2 M genetic variants (the proportion of causal genetic variants for CRC assumed to be 0.003), whereas the PRS of the 140 known variants identified from GWASs had the lowest AUC (AUC = 0.629). Based on the LDpred-derived PRS, we are able to identify 30% of individuals without a family history as having risk for CRC similar to those with a family history of CRC, whereas the PRS based on known GWAS variants identified only top 10% as having a similar relative risk. About 90% of these individuals have no family history and would have been considered average risk under current screening guidelines, but might benefit from earlier screening. The developed PRS offers a way for risk-stratified CRC screening and other targeted interventions.
Keywords: cancer risk prediction; colorectal cancer; machine learning; polygenic risk score.
Copyright © 2020 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
Age dependency of the polygenic risk score for colorectal cancer.Am J Hum Genet. 2021 Mar 4;108(3):525-526. doi: 10.1016/j.ajhg.2021.02.002. Am J Hum Genet. 2021. PMID: 33667395 Free PMC article. No abstract available.
-
Response to Li and Hopper.Am J Hum Genet. 2021 Mar 4;108(3):527-529. doi: 10.1016/j.ajhg.2021.02.003. Am J Hum Genet. 2021. PMID: 33667396 Free PMC article. No abstract available.
References
-
- Howlader N., Noone A.M., Krapcho M., Miller D. National Cancer Institute; Bethesda, MD: 2019. SEER Cancer Statistics Review, 1975-2016.https://seer.cancer.gov/archive/csr/1975_2016/
-
- Vogelaar I., van Ballegooijen M., Schrag D., Boer R., Winawer S.J., Habbema J.D.F., Zauber A.G. How much can current interventions reduce colorectal cancer mortality in the U.S.? Mortality projections for scenarios of risk-factor modification, screening, and treatment. Cancer. 2006;107:1624–1633. - PubMed
-
- Smith R.A., Mettlin C.J., Davis K.J., Eyre H. American Cancer Society guidelines for the early detection of cancer. CA Cancer J. Clin. 2000;50:34–49. - PubMed
MeSH terms
Grants and funding
- P30 ES010126/ES/NIEHS NIH HHS/United States
- U10 CA037429/CA/NCI NIH HHS/United States
- R01 CA195789/CA/NCI NIH HHS/United States
- U01 CA137088/CA/NCI NIH HHS/United States
- R01 CA223498/CA/NCI NIH HHS/United States
- UM1 CA186107/CA/NCI NIH HHS/United States
- 24390/CRUK_/Cancer Research UK/United Kingdom
- 10589/CRUK_/Cancer Research UK/United Kingdom
- 001/WHO_/World Health Organization/International
- R01 CA206279/CA/NCI NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- UG1 CA189974/CA/NCI NIH HHS/United States
- T32 CA163177/CA/NCI NIH HHS/United States
- U01 CA206110/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- 19167/CRUK_/Cancer Research UK/United Kingdom
- U01 CA167551/CA/NCI NIH HHS/United States
- P20 CA252728/CA/NCI NIH HHS/United States
- K07 CA188142/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

