Symbiosis between nanohaloarchaeon and haloarchaeon is based on utilization of different polysaccharides
- PMID: 32759215
- PMCID: PMC7443923
- DOI: 10.1073/pnas.2007232117
Symbiosis between nanohaloarchaeon and haloarchaeon is based on utilization of different polysaccharides
Abstract
Nano-sized archaeota, with their small genomes and limited metabolic capabilities, are known to associate with other microbes, thereby compensating for their own auxotrophies. These diminutive and yet ubiquitous organisms thrive in hypersaline habitats that they share with haloarchaea. Here, we reveal the genetic and physiological nature of a nanohaloarchaeon-haloarchaeon association, with both microbes obtained from a solar saltern and reproducibly cultivated together in vitro. The nanohaloarchaeon Candidatus Nanohalobium constans LC1Nh is an aerotolerant, sugar-fermenting anaerobe, lacking key anabolic machinery and respiratory complexes. The nanohaloarchaeon cells are found physically connected to the chitinolytic haloarchaeon Halomicrobium sp. LC1Hm. Our experiments revealed that this haloarchaeon can hydrolyze chitin outside the cell (to produce the monosaccharide N-acetylglucosamine), using this beta-glucan to obtain carbon and energy for growth. However, LC1Hm could not metabolize either glycogen or starch (both alpha-glucans) or other polysaccharides tested. Remarkably, the nanohaloarchaeon's ability to hydrolyze glycogen and starch to glucose enabled growth of Halomicrobium sp. LC1Hm in the absence of a chitin. These findings indicated that the nanohaloarchaeon-haloarchaeon association is both mutualistic and symbiotic; in this case, each microbe relies on its partner's ability to degrade different polysaccharides. This suggests, in turn, that other nano-sized archaeota may also be beneficial for their hosts. Given that availability of carbon substrates can vary both spatially and temporarily, the susceptibility of Halomicrobium to colonization by Ca Nanohalobium can be interpreted as a strategy to maximize the long-term fitness of the host.
Keywords: haloarchaea; nanohaloarchaea; polysaccharide utilization; solar salterns; symbiosis.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
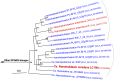

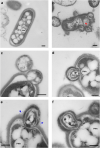



Similar articles
-
Nanohaloarchaea as beneficiaries of xylan degradation by haloarchaea.Microb Biotechnol. 2023 Sep;16(9):1803-1822. doi: 10.1111/1751-7915.14272. Epub 2023 Jun 14. Microb Biotechnol. 2023. PMID: 37317055 Free PMC article.
-
Single-cell genomics of co-sorted Nanoarchaeota suggests novel putative host associations and diversification of proteins involved in symbiosis.Microbiome. 2018 Sep 17;6(1):161. doi: 10.1186/s40168-018-0539-8. Microbiome. 2018. PMID: 30223889 Free PMC article.
-
Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park.Biol Direct. 2013 Apr 22;8:9. doi: 10.1186/1745-6150-8-9. Biol Direct. 2013. PMID: 23607440 Free PMC article.
-
From genomes to function: haloarchaea as model organisms.Microbiology (Reading). 2006 Mar;152(Pt 3):585-590. doi: 10.1099/mic.0.28504-0. Microbiology (Reading). 2006. PMID: 16514139 Review.
-
Happy together: genomic insights into the unique Nanoarchaeum/Ignicoccus association.J Biol. 2009;8(1):7. doi: 10.1186/jbiol110. Epub 2009 Jan 23. J Biol. 2009. PMID: 19216728 Free PMC article. Review.
Cited by
-
Architecture, Function, Regulation, and Evolution of α-Glucans Metabolic Enzymes in Prokaryotes.Chem Rev. 2024 Apr 24;124(8):4863-4934. doi: 10.1021/acs.chemrev.3c00811. Epub 2024 Apr 12. Chem Rev. 2024. PMID: 38606812 Free PMC article. Review.
-
The cell biology of archaea.Nat Microbiol. 2022 Nov;7(11):1744-1755. doi: 10.1038/s41564-022-01215-8. Epub 2022 Oct 17. Nat Microbiol. 2022. PMID: 36253512 Free PMC article. Review.
-
Metagenomic characterization of viruses and mobile genetic elements associated with the DPANN archaeal superphylum.Nat Microbiol. 2024 Dec;9(12):3362-3375. doi: 10.1038/s41564-024-01839-y. Epub 2024 Oct 24. Nat Microbiol. 2024. PMID: 39448846
-
Deciphering Symbiotic Interactions of "Candidatus Aenigmarchaeota" with Inferred Horizontal Gene Transfers and Co-occurrence Networks.mSystems. 2021 Aug 31;6(4):e0060621. doi: 10.1128/mSystems.00606-21. Epub 2021 Jul 27. mSystems. 2021. PMID: 34313464 Free PMC article.
-
Distinct life cycle stages of an ectosymbiotic DPANN archaeon.ISME J. 2024 Jan 8;18(1):wrae076. doi: 10.1093/ismejo/wrae076. ISME J. 2024. PMID: 38691426 Free PMC article.
References
-
- Lee C. J. D. et al. ., NaCl-saturated brines are thermodynamically moderate, rather than extreme, microbial habitats. FEMS Microbiol. Rev. 42, 672–693 (2018). - PubMed
-
- Collins N. C., Population ecology of Ephydra cinerea Jones (Diptera: Ephydridae), the only benthic metazoan of the Great Salt Lake, U.S.A. Hydrobiologia 68, 99–112 (1980).
-
- Zmora O., Avital E., Gordin H., Results of an attempt for mass production of Artemia in extensive ponds. Aquaculture 213, 395–400 (2002).
-
- Litvinenko L. I., Litvinenko A. I., Boiko E. G., Kutsanov K., Artemia cyst production in Russia. Chin. J. Oceanology Limnol. 33, 1436–1450 (2015).
-
- Korovessis N. A., Lekkas T. D., Solar Saltworks’ wetland function. Glob. NEST J. 11, 49–57 (2009).

