Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary
- PMID: 32759216
- PMCID: PMC7443898
- DOI: 10.1073/pnas.2005570117
Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary
Abstract
We sequenced more than 52,500 single cells from embryonic day 11.5 (E11.5) postembryonic day 5 (P5) gonads and performed lineage tracing to analyze primordial follicles and wave 1 medullar follicles during mouse fetal and perinatal oogenesis. Germ cells clustered into six meiotic substages, as well as dying/nurse cells. Wnt-expressing bipotential precursors already present at E11.5 are followed at each developmental stage by two groups of ovarian pregranulosa (PG) cells. One PG group, bipotential pregranulosa (BPG) cells, derives directly from bipotential precursors, expresses Foxl2 early, and associates with cysts throughout the ovary by E12.5. A second PG group, epithelial pregranulosa (EPG) cells, arises in the ovarian surface epithelium, ingresses cortically by E12.5 or earlier, expresses Lgr5, but delays robust Foxl2 expression until after birth. By E19.5, EPG cells predominate in the cortex and differentiate into granulosa cells of quiescent primordial follicles. In contrast, medullar BPG cells differentiate along a distinct pathway to become wave 1 granulosa cells. Reflecting their separate somatic cellular lineages, second wave follicles were ablated by diptheria toxin treatment of Lgr5-DTR-EGFP mice at E16.5 while first wave follicles developed normally and supported fertility. These studies provide insights into ovarian somatic cells and a resource to study the development, physiology, and evolutionary conservation of mammalian ovarian follicles.
Keywords: follicle; mouse; ovary; pregranulosa; scRNAseq.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


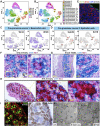

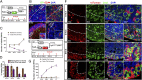

References
-
- Hirshfield A. N., Development of follicles in the mammalian ovary. Int. Rev. Cytol. 124, 43–101 (1991). - PubMed
-
- McLaren A., Development of the mammalian gonad: The fate of the supporting cell lineage. BioEssays 13, 151–156 (1991). - PubMed
-
- Pepling M. E., Spradling A. C., Female mouse germ cells form synchronously dividing cysts. Development 125, 3323–3328 (1998). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

