Patient ancestry significantly contributes to molecular heterogeneity of systemic lupus erythematosus
- PMID: 32759501
- PMCID: PMC7455079
- DOI: 10.1172/jci.insight.140380
Patient ancestry significantly contributes to molecular heterogeneity of systemic lupus erythematosus
Abstract
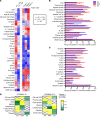
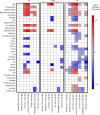
Gene expression signatures can stratify patients with heterogeneous diseases, such as systemic lupus erythematosus (SLE), yet understanding the contributions of ancestral background to this heterogeneity is not well understood. We hypothesized that ancestry would significantly influence gene expression signatures and measured 34 gene modules in 1566 SLE patients of African ancestry (AA), European ancestry (EA), or Native American ancestry (NAA). Healthy subject ancestry-specific gene expression provided the transcriptomic background upon which the SLE patient signatures were built. Although standard therapy affected every gene signature and significantly increased myeloid cell signatures, logistic regression analysis determined that ancestral background significantly changed 23 of 34 gene signatures. Additionally, the strongest association to gene expression changes was found with autoantibodies, and this also had etiology in ancestry: the AA predisposition to have both RNP and dsDNA autoantibodies compared with EA predisposition to have only anti-dsDNA. A machine learning approach was used to determine a gene signature characteristic to distinguish AA SLE and was most influenced by genes characteristic of the perturbed B cell axis in AA SLE patients.
Keywords: Autoimmunity; Rheumatology.
Conflict of interest statement
Figures





Similar articles
-
IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus.Ann Rheum Dis. 2012 Mar;71(3):463-8. doi: 10.1136/annrheumdis-2011-200463. Epub 2011 Nov 16. Ann Rheum Dis. 2012. PMID: 22088620 Free PMC article.
-
PTPN22 association in systemic lupus erythematosus (SLE) with respect to individual ancestry and clinical sub-phenotypes.PLoS One. 2013 Aug 7;8(8):e69404. doi: 10.1371/journal.pone.0069404. eCollection 2013. PLoS One. 2013. PMID: 23950893 Free PMC article.
-
APOL1 and the risk of adverse renal outcomes in patients of African ancestry with systemic lupus erythematosus.Lupus. 2023 May;32(6):763-770. doi: 10.1177/09612033231172660. Epub 2023 Apr 27. Lupus. 2023. PMID: 37105192 Free PMC article.
-
The pathogenesis of systemic lupus erythematosus: Harnessing big data to understand the molecular basis of lupus.J Autoimmun. 2020 Jun;110:102359. doi: 10.1016/j.jaut.2019.102359. Epub 2019 Dec 2. J Autoimmun. 2020. PMID: 31806421 Review.
-
A systematic literature review on the European, African and Amerindian genetic ancestry components on Brazilian health outcomes.Sci Rep. 2019 Jun 20;9(1):8874. doi: 10.1038/s41598-019-45081-7. Sci Rep. 2019. PMID: 31221977 Free PMC article.
Cited by
-
Unraveling transcriptomic signatures and dysregulated pathways in systemic lupus erythematosus across disease states.Arthritis Res Ther. 2024 May 13;26(1):99. doi: 10.1186/s13075-024-03327-4. Arthritis Res Ther. 2024. PMID: 38741185 Free PMC article.
-
Differential regulation of the interferon response in systemic lupus erythematosus distinguishes patients of Asian ancestry.RMD Open. 2023 Sep;9(3):e003475. doi: 10.1136/rmdopen-2023-003475. RMD Open. 2023. PMID: 37709528 Free PMC article.
-
An interpretable machine learning pipeline based on transcriptomics predicts phenotypes of lupus patients.iScience. 2023 Sep 25;26(10):108042. doi: 10.1016/j.isci.2023.108042. eCollection 2023 Oct 20. iScience. 2023. PMID: 37860757 Free PMC article.
-
Sub-setting systemic lupus erythematosus by combined molecular phenotypes defines divergent populations in two phase III randomized trials.Rheumatology (Oxford). 2021 Nov 3;60(11):5390-5396. doi: 10.1093/rheumatology/keab144. Rheumatology (Oxford). 2021. PMID: 33580248 Free PMC article. Clinical Trial.
-
IFN-associated B cell hyperactivity is highly enriched in SLE patients harboring Sm/RNP antibodies.bioRxiv [Preprint]. 2024 Nov 19:2024.11.18.624119. doi: 10.1101/2024.11.18.624119. bioRxiv. 2024. PMID: 39605660 Free PMC article. Preprint.
References
-
- Ferretti C, La Cava A. Overview of the pathogenesis of systemic lupus erythematosus. In: Tsokos GC, ed. Systemic lupus erythematosus—basic, applied and clinical aspects. Academic Press; 2016:55–62.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

