Loss of Sbds in zebrafish leads to neutropenia and pancreas and liver atrophy
- PMID: 32759502
- PMCID: PMC7526460
- DOI: 10.1172/jci.insight.134309
Loss of Sbds in zebrafish leads to neutropenia and pancreas and liver atrophy
Abstract
Shwachman-Diamond syndrome (SDS) is characterized by exocrine pancreatic insufficiency, neutropenia, and skeletal abnormalities. Biallelic mutations in SBDS, which encodes a ribosome maturation factor, are found in 90% of SDS cases. Sbds-/- mice are embryonic lethal. Using CRISPR/Cas9 editing, we created sbds-deficient zebrafish strains. Sbds protein levels progressively decreased and became undetectable at 10 days postfertilization (dpf). Polysome analysis revealed decreased 80S ribosomes. Homozygous mutant fish developed normally until 15 dpf. Mutant fish subsequently had stunted growth and showed signs of atrophy in pancreas, liver, and intestine. In addition, neutropenia occurred by 5 dpf. Upregulation of tp53 mRNA did not occur until 10 dpf, and inhibition of proliferation correlated with death by 21 dpf. Transcriptome analysis showed tp53 activation through upregulation of genes involved in cell cycle arrest, cdkn1a and ccng1, and apoptosis, puma and mdm2. However, elimination of Tp53 function did not prevent lethality. Because of growth retardation and atrophy of intestinal epithelia, we studied the effects of starvation on WT fish. Starved WT fish showed intestinal atrophy, zymogen granule loss, and tp53 upregulation - similar to the mutant phenotype. In addition, there was reduction in neutral lipid storage and ribosomal protein amount, similar to the mutant phenotype. Thus, loss of Sbds in zebrafish phenocopies much of the human disease and is associated with growth arrest and tissue atrophy, particularly of the gastrointestinal system, at the larval stage. A variety of stress responses, some associated with Tp53, contribute to pathophysiology of SDS.
Keywords: Embryonic development; Gastroenterology; Genetic diseases; Hematology; P53.
Conflict of interest statement
Figures

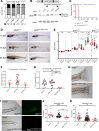

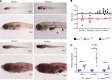

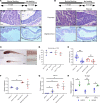
References
-
- Rothbaum R, et al. Shwachman-Diamond syndrome: report from an international conference. J Pediatr. 2002;141(2):266–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

