Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists
- PMID: 32759979
- PMCID: PMC7897578
- DOI: 10.1038/s41379-020-0640-y
Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists
Abstract
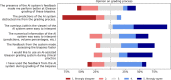
The Gleason score is the most important prognostic marker for prostate cancer patients, but it suffers from significant observer variability. Artificial intelligence (AI) systems based on deep learning can achieve pathologist-level performance at Gleason grading. However, the performance of such systems can degrade in the presence of artifacts, foreign tissue, or other anomalies. Pathologists integrating their expertise with feedback from an AI system could result in a synergy that outperforms both the individual pathologist and the system. Despite the hype around AI assistance, existing literature on this topic within the pathology domain is limited. We investigated the value of AI assistance for grading prostate biopsies. A panel of 14 observers graded 160 biopsies with and without AI assistance. Using AI, the agreement of the panel with an expert reference standard increased significantly (quadratically weighted Cohen's kappa, 0.799 vs. 0.872; p = 0.019). On an external validation set of 87 cases, the panel showed a significant increase in agreement with a panel of international experts in prostate pathology (quadratically weighted Cohen's kappa, 0.733 vs. 0.786; p = 0.003). In both experiments, on a group-level, AI-assisted pathologists outperformed the unassisted pathologists and the standalone AI system. Our results show the potential of AI systems for Gleason grading, but more importantly, show the benefits of pathologist-AI synergy.
Conflict of interest statement
WB reports grants from the Dutch Cancer Society, during the conduct of the study. JvdL reports personal fees from Philips, grants from Philips, personal fees from ContextVision, personal fees from AbbVie, grants from Sectra, outside the submitted work. GL reports grants from the Dutch Cancer Society, during the conduct of the study; grants from Philips Digital Pathology Solutions, personal fees from Novartis, outside the submitted work. MB, JAB, AB, AC, LE, ME, XF, KG, GP, PR, GS, PS, JT, HvB, RV, and CH-vdK have nothing to disclose.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

