Molecular Characterization and Event-Specific Real-Time PCR Detection of Two Dissimilar Groups of Genetically Modified Petunia (Petunia x hybrida) Sold on the Market
- PMID: 32760413
- PMCID: PMC7372090
- DOI: 10.3389/fpls.2020.01047
Molecular Characterization and Event-Specific Real-Time PCR Detection of Two Dissimilar Groups of Genetically Modified Petunia (Petunia x hybrida) Sold on the Market
Abstract
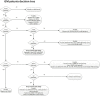
Petunia plants with unusual orange flowers were noticed on the European market and confirmed to be genetically modified (GM) by the Finnish authorities in spring 2017. Later in 2017, inspections and controls performed by several official laboratories of national competent authorities in the European Union detected several GM petunia varieties with orange flowers, but also another group of unusually colored flowers. In the latter group, a so far undetected gene coding for a flavonoid 3'5' hydroxylase (F3'5'H) responsible for the purple color was identified by German and Dutch authorities, suggesting that the petunias found on the markets contain different genetic constructs. Here, a strategy is described for the identification of GM petunia varieties. It is based on an initial GMO screening for known elements using (real-time) PCR and subsequent identification of the insertion sites by a gene walking-like approach called ALF (amplification of linearly-enriched fragments) in combination with Sanger and MinION sequencing. The results indicate that the positively identified GM petunias can be traced back to two dissimilar GM events used for breeding of the different varieties. The test results also confirm that the transgenic petunia event RL01-17 used in the first German field trial in 1991 is not the origin of the GM petunias sold on the market. On basis of the obtained sequence data, event-specific real-time PCR confirmatory methods were developed and validated. These methods are applicable for the rapid detection and identification of GM petunias in routine analysis. In addition, a decision support system was developed for revealing the most likely origin of the GM petunia.
Keywords: MinION sequencing; amplification of linearly-enriched fragments; event-specific detection; genetically modified (GM); petunia; real-time PCR; unauthorized GMO.
Copyright © 2020 Voorhuijzen, Prins, Belter, Bendiek, Brünen-Nieweler, van Dijk, Goerlich, Kok, Pickel, Scholtens, Stolz and Grohmann.
Figures




References
-
- Debode F., Janssen E., Berben G. (2013). Development of 10 new screening PCR assays for GMO detection targeting promoters (pFMV, pNOS, pSSuAra, pTA29, pUbi, pRice actin) and terminators (t35S, tE9, tOCS, tg7). Eur. Food Res. Technol. 236 (4), 659–669. 10.1007/s00217-013-1921-1 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

