Advancing biodiversity assessments with environmental DNA: Long-read technologies help reveal the drivers of Amazonian fungal diversity
- PMID: 32760545
- PMCID: PMC7391351
- DOI: 10.1002/ece3.6477
Advancing biodiversity assessments with environmental DNA: Long-read technologies help reveal the drivers of Amazonian fungal diversity
Abstract
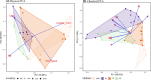
Fungi are a key component of tropical biodiversity. However, due to their inconspicuous and largely subterranean nature, they are usually neglected in biodiversity inventories. The goal of this study was to identify the key determinants of fungal richness, community composition, and turnover in tropical rainforests. We tested specifically for the effect of soil properties, habitat, and locality in Amazonia. For these analyses, we used high-throughput sequencing data of short and long reads of fungal DNA present in soil and organic litter samples, combining existing and novel genomic data. Habitat type (phytophysiognomy) emerges as the strongest factor explaining fungal community composition. Naturally open areas-campinas-are the richest habitat overall. Soil properties have different effects depending on the soil layer (litter or mineral soil) and the choice of genetic marker. We suggest that campinas could be a neglected hotspot of fungal diversity. An underlying cause for their rich diversity may be the overall low soil fertility, which increases the reliance on biotic interactions essential for nutrient absorption in these environments, notably ectomycorrhizal fungi-plant associations. Our results highlight the advantages of using both short and long DNA reads produced through high-throughput sequencing to characterize fungal diversity. While short reads can suffice for diversity and community comparison, long reads add taxonomic precision and have the potential to reveal population diversity.
Keywords: PacBio; environmental DNA; high‐throughput sequencing; metabarcoding; neotropical biodiversity; third‐generation sequencing.
© 2020 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Abarenkov, K. , Henrik Nilsson, R. , Larsson, K. H. , Alexander, I. J. , Eberhardt, U. , Erland, S. , … Sen, R. (2010). The UNITE database for molecular identification of fungi – Recent updates and future perspectives. New Phytologist, 186, 281–285. - PubMed
-
- Adeney, J. M. , Christensen, N. L. , Vicentini, A. , & Cohn‐Haft, M. (2016). White‐sand ecosystems in Amazonia. Biotropica, 48, 7–23.
-
- Arbizu, M. , & PairwiseAdonis, P. (2020). Pairwise Multilevel Comparison Using adonis. R Packag. Version 0.4.
LinkOut - more resources
Full Text Sources

