The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation
- PMID: 32762776
- PMCID: PMC7412812
- DOI: 10.1186/s13059-020-02115-y
The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation
Abstract
Background: RNA-binding proteins (RBPs) function as master regulators of gene expression. Alterations in RBP expression and function are often observed in cancer and influence critical pathways implicated in tumor initiation and growth. Identification and characterization of oncogenic RBPs and their regulatory networks provide new opportunities for targeted therapy.
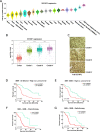
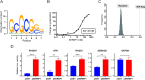
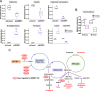
Results: We identify the RNA-binding protein SERBP1 as a novel regulator of glioblastoma (GBM) development. High SERBP1 expression is prevalent in GBMs and correlates with poor patient survival and poor response to chemo- and radiotherapy. SERBP1 knockdown causes delay in tumor growth and impacts cancer-relevant phenotypes in GBM and glioma stem cell lines. RNAcompete identifies a GC-rich region as SERBP1-binding motif; subsequent genomic and functional analyses establish SERBP1 regulation role in metabolic routes preferentially used by cancer cells. An important consequence of these functions is SERBP1 impact on methionine production. SERBP1 knockdown decreases methionine levels causing a subsequent reduction in histone methylation as shown for H3K27me3 and upregulation of genes associated with neurogenesis, neuronal differentiation, and function. Further analysis demonstrates that several of these genes are downregulated in GBM, potentially through epigenetic silencing as indicated by the presence of H3K27me3 sites.
Conclusions: SERBP1 is the first example of an RNA-binding protein functioning as a central regulator of cancer metabolism and indirect modulator of epigenetic regulation in GBM. By bridging these two processes, SERBP1 enhances glioma stem cell phenotypes and contributes to GBM poorly differentiated state.
Keywords: Cancer metabolism; Epigenetic regulation; GBM; One carbon cycle; RNA-binding protein; SERBP1.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

