Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling
- PMID: 32762848
- PMCID: PMC7442489
- DOI: 10.7554/eLife.58464
Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling
Abstract
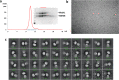
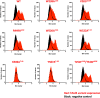
Frizzleds (Fzd) are the primary receptors for Wnt morphogens, which are essential regulators of stem cell biology, yet the structural basis of Wnt signaling through Fzd remains poorly understood. Here we report the structure of an unliganded human Fzd5 determined by single-particle cryo-EM at 3.7 Å resolution, with the aid of an antibody chaperone acting as a fiducial marker. We also analyzed the topology of low-resolution XWnt8/Fzd5 complex particles, which revealed extreme flexibility between the Wnt/Fzd-CRD and the Fzd-TM regions. Analysis of Wnt/β-catenin signaling in response to Wnt3a versus a 'surrogate agonist' that cross-links Fzd to LRP6, revealed identical structure-activity relationships. Thus, canonical Wnt/β-catenin signaling appears to be principally reliant on ligand-induced Fzd/LRP6 heterodimerization, versus the allosteric mechanisms seen in structurally analogous class A G protein-coupled receptors, and Smoothened. These findings deepen our mechanistic understanding of Wnt signal transduction, and have implications for harnessing Wnt agonism in regenerative medicine.
Keywords: Cryo-EM; Frizzled; Wnt; canonical Wnt/β-catenin signaling; human; molecular biophysics; structural biology; surrogate Wnt agonist.
© 2020, Tsutsumi et al.
Conflict of interest statement
NT, SM, DW, KJ, YM, JB, NA, AK, CG No competing interests declared, CJ, KG KCG and CYJ are founders of Surrozen Therapeutics.
Figures








References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Audet M, White KL, Breton B, Zarzycka B, Han GW, Lu Y, Gati C, Batyuk A, Popov P, Velasquez J, Manahan D, Hu H, Weierstall U, Liu W, Shui W, Katritch V, Cherezov V, Hanson MA, Stevens RC. Crystal structure of misoprostol bound to the labor inducer prostaglandin E2 receptor. Nature Chemical Biology. 2019;15:11–17. doi: 10.1038/s41589-018-0160-y. - DOI - PMC - PubMed
-
- Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, Sansom MSP, Newstead S, Rohatgi R, Siebold C. Structural basis of smoothened regulation by its extracellular domains. Nature. 2016;535:517–522. doi: 10.1038/nature18934. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

