Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front
- PMID: 32763154
- PMCID: PMC7479520
- DOI: 10.1016/j.cell.2020.07.005
Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front
Erratum in
-
Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front.Cell. 2020 Oct 29;183(3):838. doi: 10.1016/j.cell.2020.10.021. Cell. 2020. PMID: 33125896 Free PMC article. No abstract available.
Abstract
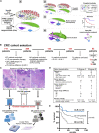
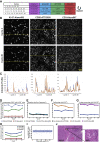
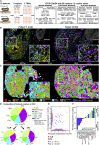
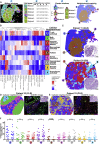
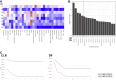
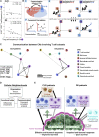
Antitumoral immunity requires organized, spatially nuanced interactions between components of the immune tumor microenvironment (iTME). Understanding this coordinated behavior in effective versus ineffective tumor control will advance immunotherapies. We re-engineered co-detection by indexing (CODEX) for paraffin-embedded tissue microarrays, enabling simultaneous profiling of 140 tissue regions from 35 advanced-stage colorectal cancer (CRC) patients with 56 protein markers. We identified nine conserved, distinct cellular neighborhoods (CNs)-a collection of components characteristic of the CRC iTME. Enrichment of PD-1+CD4+ T cells only within a granulocyte CN positively correlated with survival in a high-risk patient subset. Coupling of tumor and immune CNs, fragmentation of T cell and macrophage CNs, and disruption of inter-CN communication was associated with inferior outcomes. This study provides a framework for interrogating how complex biological processes, such as antitumoral immunity, occur through concerted actions of cells and spatial domains.
Keywords: CODEX; FFPE; antitumoral immunity; cellular neighborhoods; colorectal cancer; immune checkpoints; immune tumor microenvironment; multiplexed imaging; tertiary lymphoid structures; tissue architecture.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests G.P.N. has received research grants from Pfizer, Vaxart, Celgene, and Juno Therapeutics during the course of this work. G.P.N., Y.G., and N.S. have equity in and are scientific advisory board members of Akoya Biosciences. C.M.S. is a scientific advisor to Enable Medicine. Akoya Biosciences makes reagents and instruments that are dependent on licenses from Stanford University. Stanford University has been granted US patent 9909167, which covers some aspects of the technology described in this paper.
Figures








References
-
- Ali H.R., Jackson H.W., Zanotelli V.R.T., Danenberg E., Fischer J.R., Bardwell H., Provenzano E., Ali H.R., Al Sa’d M., Alon S. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer. 2020;1:163–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UG3 DK114937/DK/NIDDK NIH HHS/United States
- U19 AI100627/AI/NIAID NIH HHS/United States
- R01 HL120724/HL/NHLBI NIH HHS/United States
- U01 AI101984/AI/NIAID NIH HHS/United States
- U54 HG010426/HG/NHGRI NIH HHS/United States
- C27165/A29073/CRUK_/Cancer Research UK/United Kingdom
- F99 CA212231/CA/NCI NIH HHS/United States
- U01 AI140498/AI/NIAID NIH HHS/United States
- U19 AI135976/AI/NIAID NIH HHS/United States
- P01 AI131374/AI/NIAID NIH HHS/United States
- 27145/CRUK_/Cancer Research UK/United Kingdom
- T32 AI007290/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U2C CA233238/CA/NCI NIH HHS/United States
- F32 CA233203/CA/NCI NIH HHS/United States
- P01 HL108797/HL/NHLBI NIH HHS/United States
- U2C CA233195/CA/NCI NIH HHS/United States
- R01 HL128173/HL/NHLBI NIH HHS/United States
- R33 CA183692/CA/NCI NIH HHS/United States
- T32 AR007422/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

