A molecular pore spans the double membrane of the coronavirus replication organelle
- PMID: 32763915
- PMCID: PMC7665310
- DOI: 10.1126/science.abd3629
A molecular pore spans the double membrane of the coronavirus replication organelle
Abstract
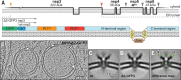
Coronavirus genome replication is associated with virus-induced cytosolic double-membrane vesicles, which may provide a tailored microenvironment for viral RNA synthesis in the infected cell. However, it is unclear how newly synthesized genomes and messenger RNAs can travel from these sealed replication compartments to the cytosol to ensure their translation and the assembly of progeny virions. In this study, we used cellular cryo-electron microscopy to visualize a molecular pore complex that spans both membranes of the double-membrane vesicle and would allow export of RNA to the cytosol. A hexameric assembly of a large viral transmembrane protein was found to form the core of the crown-shaped complex. This coronavirus-specific structure likely plays a key role in coronavirus replication and thus constitutes a potential drug target.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



Comment in
-
Coronavirus dons a new crown.Science. 2020 Sep 11;369(6509):1306-1307. doi: 10.1126/science.abe0322. Science. 2020. PMID: 32913092 No abstract available.
References
-
- Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., Rollin P. E., Dowell S. F., Ling A. E., Humphrey C. D., Shieh W. J., Guarner J., Paddock C. D., Rota P., Fields B., DeRisi J., Yang J. Y., Cox N., Hughes J. M., LeDuc J. W., Bellini W. J., Anderson L. J., Group S. W.; SARS Working Group , A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003). 10.1056/NEJMoa030781 - DOI - PubMed
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., China Novel Coronavirus Investigating and Research Team, A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J., SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLOS Biol. 6, e226 (2008). 10.1371/journal.pbio.0060226 - DOI - PMC - PubMed
-
- Snijder E. J., Limpens R. W. A. L., de Wilde A. H., de Jong A. W. M., Zevenhoven-Dobbe J. C., Maier H. J., Faas F. F. G. A., Koster A. J., Bárcena M., A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLOS Biol. 18, e3000715 (2020). 10.1371/journal.pbio.3000715 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

