KAT7 is a genetic vulnerability of acute myeloid leukemias driven by MLL rearrangements
- PMID: 32764680
- PMCID: PMC7610570
- DOI: 10.1038/s41375-020-1001-z
KAT7 is a genetic vulnerability of acute myeloid leukemias driven by MLL rearrangements
Abstract
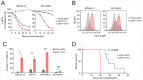
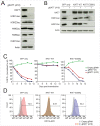
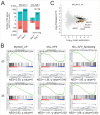
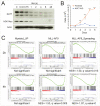
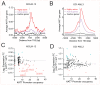
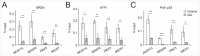
Histone acetyltransferases (HATs) catalyze the transfer of an acetyl group from acetyl-CoA to lysine residues of histones and play a central role in transcriptional regulation in diverse biological processes. Dysregulation of HAT activity can lead to human diseases including developmental disorders and cancer. Through genome-wide CRISPR-Cas9 screens, we identified several HATs of the MYST family as fitness genes for acute myeloid leukemia (AML). Here we investigate the essentiality of lysine acetyltransferase KAT7 in AMLs driven by the MLL-X gene fusions. We found that KAT7 loss leads to a rapid and complete loss of both H3K14ac and H4K12ac marks, in association with reduced proliferation, increased apoptosis, and differentiation of AML cells. Acetyltransferase activity of KAT7 is essential for the proliferation of these cells. Mechanistically, our data propose that acetylated histones provide a platform for the recruitment of MLL-fusion-associated adaptor proteins such as BRD4 and AF4 to gene promoters. Upon KAT7 loss, these factors together with RNA polymerase II rapidly dissociate from several MLL-fusion target genes that are essential for AML cell proliferation, including MEIS1, PBX3, and SENP6. Our findings reveal that KAT7 is a plausible therapeutic target for this poor prognosis AML subtype.
Conflict of interest statement
G.S.V. is a consultant for Kymab and Oxstem.
Figures
References
-
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

