Identification of chemosensory genes from the antennal transcriptome of Semiothisa cinerearia
- PMID: 32764791
- PMCID: PMC7413487
- DOI: 10.1371/journal.pone.0237134
Identification of chemosensory genes from the antennal transcriptome of Semiothisa cinerearia
Abstract
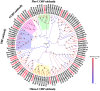
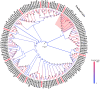
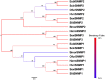
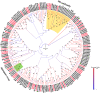
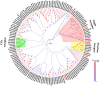
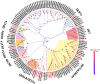
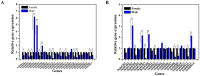
Olfaction plays vital roles in the survival and reproduction of insects. The completion of olfactory recognition requires the participation of various complex protein families. However, little is known about the olfactory-related proteins in Semiothisa cinerearia Bremer et Grey, an important pest of Chinese scholar tree. In this study, we sequenced the antennal transcriptome of S. cinerearia and identified 125 olfactory-related genes, including 25 odorant-binding proteins (OBPs), 15 chemosensory proteins (CSPs), two sensory neuron membrane proteins (SNMPs), 52 odorant receptors (ORs), eight gustatory receptors (GRs) and 23 ionotropic receptors (IRs). BLASTX best hit results and phylogenetic analyses indicated that these genes were most identical to their respective orthologs from Ectropis obliqua. Further quantitative real-time PCR (qRT-PCR) analysis revealed that three ScinOBPs and three ScinORs were highly expressed in male antennae, while seven ScinOBPs and twelve ScinORs were female-specifically expressed. Our study will be useful for the elucidation of olfactory mechanisms in S. cinerearia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Sun L, Mao TF, Zhang YX, Wu JJ, Bai JH, Zhang YN et al. Characterization of candidate odorant-binding proteins and chemosensory proteins in the tea geometrid Ectropis obliqua Prout (Lepidoptera: Geometridae). Archives of Insect Biochemistry and Physiology. 2017. April; 94(4): e21383 10.1002/arch.21383 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

