Genomic Surveillance of Yellow Fever Virus Epizootic in São Paulo, Brazil, 2016 - 2018
- PMID: 32764827
- PMCID: PMC7437926
- DOI: 10.1371/journal.ppat.1008699
Genomic Surveillance of Yellow Fever Virus Epizootic in São Paulo, Brazil, 2016 - 2018
Abstract
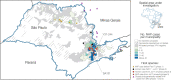
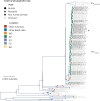
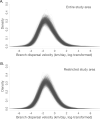
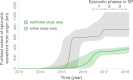
São Paulo, a densely inhabited state in southeast Brazil that contains the fourth most populated city in the world, recently experienced its largest yellow fever virus (YFV) outbreak in decades. YFV does not normally circulate extensively in São Paulo, so most people were unvaccinated when the outbreak began. Surveillance in non-human primates (NHPs) is important for determining the magnitude and geographic extent of an epizootic, thereby helping to evaluate the risk of YFV spillover to humans. Data from infected NHPs can give more accurate insights into YFV spread than when using data from human cases alone. To contextualise human cases, identify epizootic foci and uncover the rate and direction of YFV spread in São Paulo, we generated and analysed virus genomic data and epizootic case data from NHPs in São Paulo. We report the occurrence of three spatiotemporally distinct phases of the outbreak in São Paulo prior to February 2018. We generated 51 new virus genomes from YFV positive cases identified in 23 different municipalities in São Paulo, mostly sampled from NHPs between October 2016 and January 2018. Although we observe substantial heterogeneity in lineage dispersal velocities between phylogenetic branches, continuous phylogeographic analyses of generated YFV genomes suggest that YFV lineages spread in São Paulo at a mean rate of approximately 1km per day during all phases of the outbreak. Viral lineages from the first epizootic phase in northern São Paulo subsequently dispersed towards the south of the state to cause the second and third epizootic phases there. This alters our understanding of how YFV was introduced into the densely populated south of São Paulo state. Our results shed light on the sylvatic transmission of YFV in highly fragmented forested regions in São Paulo state and highlight the importance of continued surveillance of zoonotic pathogens in sentinel species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Vasconcelos PFC, Sperb AF, Monteiro HAO, Torres MAN, Sousa MRS, Vasconcelos HB, et al. Isolations of yellow fever virus from Haemagogus leucocelaenus in Rio Grande do Sul State, Brazil. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2003;97: 60–62. 10.1016/s0035-9203(03)90023-x - DOI - PubMed
-
- Abreu FVS de, Ribeiro IP, Ferreira-de-Brito A, Santos AAC dos, Miranda RM de, Bonelly I de S, et al. Haemagogus leucocelaenus and Haemagogus janthinomys are the primary vectors in the major yellow fever outbreak in Brazil, 2016–2018. Emerging Microbes & Infections. 2019;8: 218–231. 10.1080/22221751.2019.1568180 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

