Heat Shock Protein Member 8 Is an Attachment Factor for Infectious Bronchitis Virus
- PMID: 32765462
- PMCID: PMC7381282
- DOI: 10.3389/fmicb.2020.01630
Heat Shock Protein Member 8 Is an Attachment Factor for Infectious Bronchitis Virus
Abstract
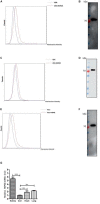
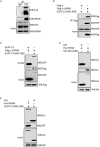
Although infectious bronchitis virus (IBV) is the first coronavirus identified, little is known about which membrane protein of host cells could interact with IBV spike protein and facilitate the infection by the virus. In this study, by using a monoclonal antibody to the S1 protein of IBV M41 strain, we found that heat shock protein member 8 (HSPA8) could interact with spike protein of IBV. HSPA8 was found to be present on the cell membrane and chicken tissues, with highest expression level in the kidney. Results of co-IP and GST-pull-down assays indicated that the receptor binding domain (RBD) of IBV M41 could interact with HSPA8. The results of binding blocking assay and infection inhibition assay showed that recombinant protein HSPA8 and antibody to HSPA8 could inhibit IBV M41 infection of chicken embryonic kidney (CEK) cells. Further, we found that HSPA8 interacted with the N-terminal 19-272 amino acids of S1 of IBV Beaudette, H120 and QX strains and HSPA8 from human and pig also interacted with IBV M41-RBD. Finally the results of binding blocking assay and infection inhibition assay showed that recombinant HSPA8 protein and antibody to HSPA8 could inhibit IBV Beaudette strain infection of Vero cells that were treated with heparanase to remove heparan sulfate from the cell surface. Taken together, our results indicate that HSPA8 is a novel host factor involved in IBV infection.
Keywords: HSPA8; attachment factor; infectious bronchitis virus; receptor binding domain; virus entry.
Copyright © 2020 Zhu, Lv, Fang, Peng, Sheng, Xiao, Kumar Ojha, Yan, Liao and Zhou.
Figures




Similar articles
-
Three Amino Acid Changes in Avian Coronavirus Spike Protein Allow Binding to Kidney Tissue.J Virol. 2020 Jan 6;94(2):e01363-19. doi: 10.1128/JVI.01363-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31694947 Free PMC article.
-
Contributions of the S2 spike ectodomain to attachment and host range of infectious bronchitis virus.Virus Res. 2013 Nov 6;177(2):127-37. doi: 10.1016/j.virusres.2013.09.006. Epub 2013 Sep 13. Virus Res. 2013. PMID: 24041648 Free PMC article.
-
The S2 Subunit of Infectious Bronchitis Virus Beaudette Is a Determinant of Cellular Tropism.J Virol. 2018 Sep 12;92(19):e01044-18. doi: 10.1128/JVI.01044-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021894 Free PMC article.
-
The avian coronavirus spike protein.Virus Res. 2014 Dec 19;194:37-48. doi: 10.1016/j.virusres.2014.10.009. Epub 2014 Oct 17. Virus Res. 2014. PMID: 25451062 Free PMC article. Review.
-
Avian infectious bronchitis virus.Rev Sci Tech. 2000 Aug;19(2):493-508. doi: 10.20506/rst.19.2.1228. Rev Sci Tech. 2000. PMID: 10935276 Review.
Cited by
-
The relation between avian coronaviruses and SARS-CoV-2 coronavirus.Front Microbiol. 2022 Oct 13;13:976462. doi: 10.3389/fmicb.2022.976462. eCollection 2022. Front Microbiol. 2022. PMID: 36312988 Free PMC article. Review.
-
Single-cell RNA-sequencing data analysis reveals a highly correlated triphasic transcriptional response to SARS-CoV-2 infection.Commun Biol. 2022 Nov 27;5(1):1302. doi: 10.1038/s42003-022-04253-4. Commun Biol. 2022. PMID: 36435849 Free PMC article.
-
Longitudinal Analysis of Biologic Correlates of COVID-19 Resolution: Case Report.Front Med (Lausanne). 2022 Jun 15;9:915367. doi: 10.3389/fmed.2022.915367. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35783607 Free PMC article.
-
Known Cellular and Receptor Interactions of Animal and Human Coronaviruses: A Review.Viruses. 2022 Feb 8;14(2):351. doi: 10.3390/v14020351. Viruses. 2022. PMID: 35215937 Free PMC article. Review.
-
In Silico Discovery and Evaluation of Inhibitors of the SARS-CoV-2 Spike Protein-HSPA8 Complex Towards Developing COVID-19 Therapeutic Drugs.Viruses. 2024 Oct 31;16(11):1726. doi: 10.3390/v16111726. Viruses. 2024. PMID: 39599841 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

