Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma
- PMID: 32765498
- PMCID: PMC7379131
- DOI: 10.3389/fimmu.2020.01402
Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma
Abstract
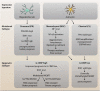
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults, with a poor prognosis, despite surgical resection combined with radio- and chemotherapy. The major clinical obstacles contributing to poor GBM prognosis are late diagnosis, diffuse infiltration, pseudo-palisading necrosis, microvascular proliferation, and resistance to conventional therapy. These challenges are further compounded by extensive inter- and intra-tumor heterogeneity and the dynamic plasticity of GBM cells. The complex heterogeneous nature of GBM cells is facilitated by the local inflammatory tumor microenvironment, which mostly induces tumor aggressiveness and drug resistance. An immunosuppressive tumor microenvironment of GBM provides multiple pathways for tumor immune evasion. Infiltrating immune cells, mostly tumor-associated macrophages, comprise much of the non-neoplastic population in GBM. Further understanding of the immune microenvironment of GBM is essential to make advances in the development of immunotherapeutics. Recently, whole-genome sequencing, epigenomics and transcriptional profiling have significantly helped improve the prognostic and therapeutic outcomes of GBM patients. Here, we discuss recent genomic advances, the role of innate and adaptive immune mechanisms, and the presence of an established immunosuppressive GBM microenvironment that suppresses and/or prevents the anti-tumor host response.
Keywords: astrocytes; brain tumor; glioblastoma; immunity; microenvironment; microglia.
Copyright © 2020 DeCordova, Shastri, Tsolaki, Yasmin, Klein, Singh and Kishore.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

