Korean Genome Project: 1094 Korean personal genomes with clinical information
- PMID: 32766443
- PMCID: PMC7385432
- DOI: 10.1126/sciadv.aaz7835
Korean Genome Project: 1094 Korean personal genomes with clinical information
Abstract
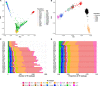
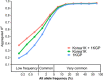
We present the initial phase of the Korean Genome Project (Korea1K), including 1094 whole genomes (sequenced at an average depth of 31×), along with data of 79 quantitative clinical traits. We identified 39 million single-nucleotide variants and indels of which half were singleton or doubleton and detected Korean-specific patterns based on several types of genomic variations. A genome-wide association study illustrated the power of whole-genome sequences for analyzing clinical traits, identifying nine more significant candidate alleles than previously reported from the same linkage disequilibrium blocks. Also, Korea1K, as a reference, showed better imputation accuracy for Koreans than the 1KGP panel. As proof of utility, germline variants in cancer samples could be filtered out more effectively when the Korea1K variome was used as a panel of normals compared to non-Korean variome sets. Overall, this study shows that Korea1K can be a useful genotypic and phenotypic resource for clinical and ethnogenetic studies.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Siska V., Jones E. R., Jeon S., Bhak Y., Kim H. M., Cho Y. S., Kim H., Lee K., Veselovskaya E., Balueva T., Gallego-Llorente M., Hofreiter M., Bradley D. G., Eriksson A., Pinhasi R., Bhak J., Manica A., Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago. Sci. Adv. 3, e1601877 (2017). - PMC - PubMed
-
- HUGO Pan-Asian SNP Consortium, Abdulla M. A., Ahmed I., Assawamakin A., Bhak J., Brahmachari S. K., Calacal G. C., Chaurasia A., Chen C. H., Chen J., Chen Y. T., Chu J., Cutiongco-de la Paz E. M., De Ungria M. C., Delfin F. C., Edo J., Fuchareon S., Ghang H., Gojobori T., Han J., Ho S. F., Hoh B. P., Huang W., Inoko H., Jha P., Jinam T. A., Jin L., Jung J., Kangwanpong D., Kampuansai J., Kennedy G. C., Khurana P., Kim H. L., Kim K., Kim S., Kim W. Y., Kimm K., Kimura R., Koike T., Kulawonganunchai S., Kumar V., Lai P. S., Lee J. Y., Lee S., Liu E. T., Majumder P. P., Mandapati K. K., Marzuki S., Mitchell W., Mukerji M., Naritomi K., Ngamphiw C., Niikawa N., Nishida N., Oh B., Oh S., Ohashi J., Oka A., Ong R., Padilla C. D., Palittapongarnpim P., Perdigon H. B., Phipps M. E., Png E., Sakaki Y., Salvador J. M., Sandraling Y., Scaria V., Seielstad M., Sidek M. R., Sinha A., Srikummool M., Sudoyo H., Sugano S., Suryadi H., Suzuki Y., Tabbada K. A., Tan A., Tokunaga K., Tongsima S., Villamor L. P., Wang E., Wang Y., Wang H., Wu J. Y., Xiao H., Xu S., Yang J. O., Shugart Y. Y., Yoo H. S., Yuan W., Zhao G., Zilfalil B. A.; Indian Genome Variation Consortium , Mapping human genetic diversity in Asia. Science 326, 1541–1545 (2009). - PubMed
-
- R. O. K. M. F. Affairs, Total Number of Overseas Koreans (2017).
-
- Databank, Population Total (2018).
-
- Seo J.-S., Rhie A., Kim J., Lee S., Sohn M.-H., Kim C.-U., Hastie A., Cao H., Yun J.-Y., Kim J., Kuk J., Park G. H., Kim J., Ryu H., Kim J., Roh M., Baek J., Hunkapiller M. W., Korlach J., Shin J.-Y., Kim C., De novo assembly and phasing of a Korean human genome. Nature 538, 243–247 (2016). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

