Identification of key genes of papillary thyroid carcinoma by integrated bioinformatics analysis
- PMID: 32766727
- PMCID: PMC7433002
- DOI: 10.1042/BSR20201555
Identification of key genes of papillary thyroid carcinoma by integrated bioinformatics analysis
Abstract
Background: Papillary thyroid carcinoma (PTC) is one of the fastest-growing malignant tumor types of thyroid cancer. Therefore, identifying the interaction of genes in PTC is crucial for elucidating its pathogenesis and finding more specific molecular biomarkers.
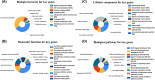
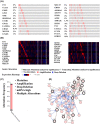
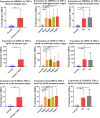
Methods: Four pairs of PTC tissues and adjacent tissues were sequenced using RNA-Seq, and 3745 differentially expressed genes were screened (P<0.05, |logFC|>1). The enrichment analysis indicated that the vast majority of differentially expressed genes (DEGs) may play a positive role in the development of cancer. Then, the significant modules were analyzed using Cytoscape software in the protein-protein interaction network. Survival analysis, TNM analysis, and immune infiltration analysis of key genes were analyzed. And the expression of ADORA1, APOE, and LPAR5 genes were verified by qPCR in PTC compared with matching adjacent tissues.
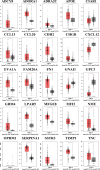
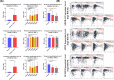
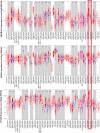
Results: Twenty-five genes were identified as hub genes with nodes greater than 10. The expression of 25 genes were verified by the GEPIA database, and the overall survival and disease-free survival analyses were conducted with Kaplan-Meier plotter. We found only three genes were confirmed with our validation and were statistically significant in PTC, namely ADORA1, APOE, and LPAR5. Further analysis found that the mRNA levels and methylation degree of these three genes were significantly correlated with the TNM staging of PTC. And these three genes were related to PTC immune infiltration. Verification of the expression of these three genes by RT-qPCR and Western blot further confirmed the reliability of our results.
Conclusion: Our study identified three genes that may play key regulatory roles in the development, metastasis, and immune infiltration of papillary thyroid carcinoma.
Keywords: RNA-Seq; bioinformatics; key gene; papillary thyroid carcinoma.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures





References
-
- Jin X., Deng B., Ye K. et al. . (2019) Comprehensive expression profiles and bioinformatics analysis reveal special circular RNA expression and potential predictability in the peripheral blood of humans with idiopathic membranous nephropathy. Mol. Med. Rep. 20, 4125–4139 10.3892/mmr.2019.10671 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

