Under the Radar: Epidemiology of Plasmodium ovale in the Democratic Republic of the Congo
- PMID: 32766832
- PMCID: PMC8006425
- DOI: 10.1093/infdis/jiaa478
Under the Radar: Epidemiology of Plasmodium ovale in the Democratic Republic of the Congo
Abstract
Background: Plasmodium ovale is an understudied malaria species prevalent throughout much of sub-Saharan Africa. Little is known about the distribution of ovale malaria and risk factors for infection in areas of high malaria endemicity.
Methods: Using the 2013 Democratic Republic of the Congo (DRC) Demographic and Health Survey, we conducted a risk factor analysis for P. ovale infections. We evaluated geographic clustering of infections and speciated to P. ovale curtisi and P. ovale wallikeri through deep sequencing.
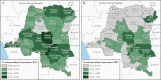
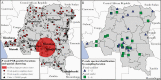
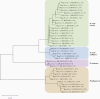
Results: Of 18 149 adults tested, we detected 143 prevalent P. ovale infections (prevalence estimate 0.8%; 95% confidence interval [CI], .59%-.98%). Prevalence ratios (PR) for significant risk factors were: male sex PR = 2.12 (95% CI, 1.38-3.26), coprevalent P. falciparum PR = 3.52 (95% CI, 2.06-5.99), and rural residence PR = 2.19 (95% CI, 1.31-3.66). P. ovale was broadly distributed throughout the DRC; an elevated cluster of infections was detected in the south-central region. Speciation revealed P. ovale curtisi and P. ovale wallikeri circulating throughout the country.
Conclusions: P. ovale persists broadly in the DRC, a high malaria burden country. For successful elimination of all malaria species, P. ovale needs to be on the radar of malaria control programs.
Keywords: Plasmodium ovale; amplicon sequencing; epidemiology; nonfalciparum malaria.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures
References
-
- Sutherland CJ, Tanomsing N, Nolder D, et al. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J Infect Dis 2010; 201:1544–50. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

