Lower cost alternatives for molecular diagnosis of COVID-19: conventional RT-PCR and SYBR Green-based RT-qPCR
- PMID: 32767275
- PMCID: PMC7411266
- DOI: 10.1007/s42770-020-00347-5
Lower cost alternatives for molecular diagnosis of COVID-19: conventional RT-PCR and SYBR Green-based RT-qPCR
Abstract
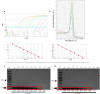
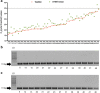
In March 2020, WHO declared a pandemic state due to SARS-CoV-2 having spread. TaqMan-based real-time RT-qPCR is currently the gold standard for COVID-19 diagnosis. However, it is a high-cost assay, inaccessible for the majority of laboratories around the world, making it difficult to diagnose on a large scale. The objective of this study was to standardize lower cost molecular methods for SARS-CoV-2 identification. E gene primers previously determined for TaqMan assays by Colman et al. (2020) were adapted in SYBR Green assay and RT-PCR conventional. The cross-reactivity test was performed with 17 positive samples for other respiratory viruses, and the sensibility test was performed with 8 dilutions (10 based) of SARS-CoV-2 isolated and 63 SARS-CoV-2-positive samples. The SYBR Green assays and conventional RT-PCR have not shown amplification of the 17 respiratory samples positives for other viruses. The SYBR Green-based assay was able to detect all 8 dilutions of the isolate. The conventional PCR detected until 107 dilution, both assays detected the majority of the 63 samples, 98.42% of positivity in SYBR Green, and 93% in conventional PCR. The average Ct variation between SYBR Green and TaqMan was 1.92 and the highest Ct detected by conventional PCR was 35.98. Both of the proposed assays are less sensitive than the current gold standard; however, our data shows a low sensibility variation, suggesting that these methods could be used by laboratories as a lower cost molecular method for SARS-CoV-2 diagnosis.
Keywords: COVID-19 pandemic; Conventional PCR; Molecular diagnoses; SARS-CoV-2; SYBR Green assay.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Automated SARS-COV-2 RNA extraction from patient nasopharyngeal samples using a modified DNA extraction kit for high throughput testing.Ann Saudi Med. 2020 Sep-Oct;40(5):373-381. doi: 10.5144/0256-4947.2020.373. Epub 2020 Oct 1. Ann Saudi Med. 2020. PMID: 32954791 Free PMC article.
-
Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19.Med Hypotheses. 2020 Aug;141:109786. doi: 10.1016/j.mehy.2020.109786. Epub 2020 Apr 25. Med Hypotheses. 2020. PMID: 32361529 Free PMC article.
-
Rapid Serological Assays and SARS-CoV-2 Real-Time Polymerase Chain Reaction Assays for the Detection of SARS-CoV-2: Comparative Study.J Med Internet Res. 2020 Oct 30;22(10):e19152. doi: 10.2196/19152. J Med Internet Res. 2020. PMID: 33031048 Free PMC article.
-
Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2.Theranostics. 2020 Jun 1;10(16):7150-7162. doi: 10.7150/thno.47649. eCollection 2020. Theranostics. 2020. PMID: 32641984 Free PMC article. Review.
-
RT-qPCR assays based on saliva rather than on nasopharyngeal swabs are possible but should be interpreted with caution: results from a systematic review and meta-analysis.Acta Biomed. 2020 Sep 7;91(3):e2020025. doi: 10.23750/abm.v91i3.10020. Acta Biomed. 2020. PMID: 32921721 Free PMC article.
Cited by
-
Current and innovative methods for the diagnosis of COVID‑19 infection (Review).Int J Mol Med. 2021 Jun;47(6):100. doi: 10.3892/ijmm.2021.4933. Epub 2021 Apr 13. Int J Mol Med. 2021. PMID: 33846767 Free PMC article. Review.
-
Rapid On-Site Detection of SARS-CoV-2 Using RT-LAMP Assay with a Portable Low-Cost Device.Biosensors (Basel). 2023 Jul 12;13(7):724. doi: 10.3390/bios13070724. Biosensors (Basel). 2023. PMID: 37504122 Free PMC article.
-
Inactivation of two SARS-CoV-2 virus surrogates by electron beam irradiation on large yellow croaker slices and their packaging surfaces.Food Control. 2023 Feb;144:109340. doi: 10.1016/j.foodcont.2022.109340. Epub 2022 Sep 6. Food Control. 2023. PMID: 36091572 Free PMC article.
-
A novel highly specific biotinylated MAC-ELISA for detection of anti-SARS-CoV-2 nucleocapsid antigen IgM antibodies during the acute phase of COVID-19.Braz J Microbiol. 2023 Dec;54(4):2893-2901. doi: 10.1007/s42770-023-01160-6. Epub 2023 Nov 6. Braz J Microbiol. 2023. PMID: 37930615 Free PMC article.
-
Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection.Clin Microbiol Rev. 2021 May 12;34(3):e00228-20. doi: 10.1128/CMR.00228-20. Print 2021 Jun 16. Clin Microbiol Rev. 2021. PMID: 33980687 Free PMC article. Review.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Organization WH (2020) Situation Report-83 HIGHLIGHTS
-
- Rodriguez-Morales AJ, Gallego V, Escalera-Antezana JP, Méndez CA, Zambrano LI, Franco-Paredes C, Suárez JA, Rodriguez-Enciso HD, Balbin-Ramon GJ, Savio-Larriera E, Risquez A, Cimerman S. COVID-19 in Latin America: the implications of the first confirmed case in Brazil. Travel Med Infect Dis. 2020;35:101613. doi: 10.1016/j.tmaid.2020.101613. - DOI - PMC - PubMed
-
- World Health Organization (2020) Laboratory testing strategy recommendations for COVID-19
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

