Alterocin, an Antibiofilm Protein Secreted by Pseudoalteromonas sp. Strain 3J6
- PMID: 32769182
- PMCID: PMC7531970
- DOI: 10.1128/AEM.00893-20
Alterocin, an Antibiofilm Protein Secreted by Pseudoalteromonas sp. Strain 3J6
Erratum in
-
Erratum for Jouault et al., "Alterocin, an Antibiofilm Protein Secreted by Pseudoalteromonas sp. Strain 3J6".Appl Environ Microbiol. 2020 Nov 10;86(23):e02431-20. doi: 10.1128/AEM.02431-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 33172901 Free PMC article. No abstract available.
Abstract
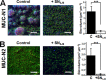
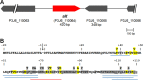
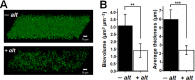
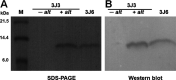
We sought to identify and study the antibiofilm protein secreted by the marine bacterium Pseudoalteromonas sp. strain 3J6. The latter is active against marine and terrestrial bacteria, including Pseudomonas aeruginosa clinical strains forming different biofilm types. Several amino acid sequences were obtained from the partially purified antibiofilm protein, named alterocin. The Pseudoalteromonas sp. 3J6 genome was sequenced, and a candidate alt gene was identified by comparing the genome-encoded proteins to the sequences from purified alterocin. Expressing the alt gene in another nonactive Pseudoalteromonas sp. strain, 3J3, demonstrated that it is responsible for the antibiofilm activity. Alterocin is a 139-residue protein that includes a predicted 20-residue signal sequence, which would be cleaved off upon export by the general secretion system. No sequence homology was found between alterocin and proteins of known functions. The alt gene is not part of an operon and adjacent genes do not seem related to alterocin production, immunity, or regulation, suggesting that these functions are not fulfilled by devoted proteins. During growth in liquid medium, the alt mRNA level peaked during the stationary phase. A single promoter was experimentally identified, and several inverted repeats could be binding sites for regulators. alt genes were found in about 30% of the Pseudoalteromonas genomes and in only a few instances of other marine bacteria of the Hahella and Paraglaciecola genera. Comparative genomics yielded the hypothesis that alt gene losses occurred within the Pseudoalteromonas genus. Overall, alterocin is a novel kind of antibiofilm protein of ecological and biotechnological interest.IMPORTANCE Biofilms are microbial communities that develop on solid surfaces or interfaces and are detrimental in a number of fields, including for example food industry, aquaculture, and medicine. In the latter, antibiotics are insufficient to clear biofilm infections, leading to chronic infections such as in the case of infection by Pseudomonas aeruginosa of the lungs of cystic fibrosis patients. Antibiofilm molecules are thus urgently needed to be used in conjunction with conventional antibiotics, as well as in other fields of application, especially if they are environmentally friendly molecules. Here, we describe alterocin, a novel antibiofilm protein secreted by a marine bacterium belonging to the Pseudoalteromonas genus, and its gene. Alterocin homologs were found in about 30% of Pseudoalteromonas strains, indicating that this new family of antibiofilm proteins likely plays an important albeit nonessential function in the biology of these bacteria. This study opens up the possibility of a variety of applications.
Keywords: Pseudoalteromonas; Pseudomonas aeruginosa; antibiofilm protein; biofilm.
Copyright © 2020 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

