Whole genome analysis reveals aneuploidies in early pregnancy loss in the horse
- PMID: 32769994
- PMCID: PMC7415156
- DOI: 10.1038/s41598-020-69967-z
Whole genome analysis reveals aneuploidies in early pregnancy loss in the horse
Abstract
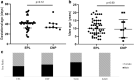
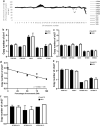
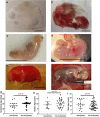
The first 8 weeks of pregnancy is a critical time, with the majority of pregnancy losses occurring during this period. Abnormal chromosome number (aneuploidy) is a common finding in human miscarriage, yet is rarely reported in domestic animals. Equine early pregnancy loss (EPL) has no diagnosis in over 80% of cases. The aim of this study was to characterise aneuploidies associated with equine EPL. Genomic DNA from clinical cases of spontaneous miscarriage (EPLs; 14-65 days of gestation) and healthy control placentae (various gestational ages) were assessed using a high density genotyping array. Aneuploidy was detected in 12/55 EPLs (21.8%), and 0/15 healthy control placentae. Whole genome sequencing (30X) and digital droplet PCR (ddPCR) validated results. The majority of these aneuploidies have never been reported in live born equines, supporting their embryonic/fetal lethality. Aneuploidies were detected in both placental and fetal compartments. Rodents are currently used to study how maternal ageing impacts aneuploidy risk, however the differences in reproductive biology is a limitation of this model. We present the first evidence of aneuploidy in naturally occurring equine EPLs at a similar rate to human miscarriage. We therefore suggest the horse as an alternative to rodent models to study mechanisms resulting in aneuploid pregnancies.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: The 'black box' of early pregnancy loss. Hum. Reprod. Update. 2002;8:333–343. - PubMed
-
- Rose BV, et al. Descriptive study of current therapeutic practices, clinical reproductive findings and incidence of pregnancy loss in intensively managed thoroughbred mares. Anim. Reprod. Sci. 2018;188:74–84. - PubMed
-
- Wiltbank MC, et al. Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology. 2016;86:239–253. - PubMed
-
- Morris L, Allen W. Reproductive efficiency of intensively managed thoroughbred mares in Newmarket. Equine Vet. J. 2002;34:51–60. - PubMed
-
- de Mestre AM, Rose BV, Chang YM, Wathes DC, Verheyen KLP. Multivariable analysis to determine risk factors associated with early pregnancy loss in thoroughbred broodmares. Theriogenology. 2019;124:18–23. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

