PIAS1 and TIF1γ collaborate to promote SnoN SUMOylation and suppression of epithelial-mesenchymal transition
- PMID: 32770107
- PMCID: PMC7853041
- DOI: 10.1038/s41418-020-0599-8
PIAS1 and TIF1γ collaborate to promote SnoN SUMOylation and suppression of epithelial-mesenchymal transition
Erratum in
-
Correction: PIAS1 and TIF1γ collaborate to promote SnoN SUMOylation and suppression of epithelial-mesenchymal transition.Cell Death Differ. 2021 May;28(5):1753. doi: 10.1038/s41418-020-00611-z. Cell Death Differ. 2021. PMID: 32811972 Free PMC article. No abstract available.
Abstract
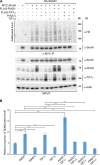
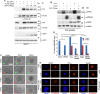
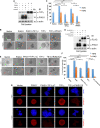
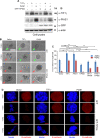
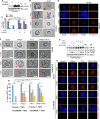
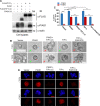
SUMO E3 ligases specify protein substrates for SUMOylation. The SUMO E3 ligases PIAS1 and TIF1γ target the transcriptional regulator SnoN for SUMOylation leading to suppression of epithelial-mesenchymal transition (EMT). Whether and how TIF1γ and PIAS1 might coordinate SnoN SUMOylation and regulation of EMT remained unknown. Here, we reveal that SnoN associates simultaneously with both TIF1γ and PIAS1, leading to a trimeric protein complex. Hence, PIAS1 and TIF1γ collaborate to promote the SUMOylation of SnoN. Importantly, loss of function studies of PIAS1 and TIF1γ suggest that these E3 ligases act in an interdependent manner to suppress EMT of breast cell-derived tissue organoids. Collectively, our findings unveil a novel mechanism by which SUMO E3 ligases coordinate substrate SUMOylation with biological implications.
Conflict of interest statement
ASC is currently employed at Fog Pharma, USA. However, this study is neither funded nor associated in any manner with Fog Pharma. The other authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

