Pyrroline-5-carboxylate synthase senses cellular stress and modulates metabolism by regulating mitochondrial respiration
- PMID: 32770108
- PMCID: PMC7853148
- DOI: 10.1038/s41418-020-0601-5
Pyrroline-5-carboxylate synthase senses cellular stress and modulates metabolism by regulating mitochondrial respiration
Abstract
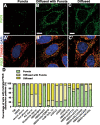
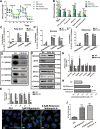
Pyrroline-5-carboxylate synthase (P5CS) catalyzes the synthesis of pyrroline-5-carboxylate (P5C), a key precursor for the synthesis of proline and ornithine. P5CS malfunction leads to multiple human diseases; however, the molecular mechanism underlying these diseases is unknown. We found that P5CS localizes in mitochondria in rod- and ring-like patterns but diffuses inside the mitochondria upon cellular starvation or exposure to oxidizing agents. Some of the human disease-related mutant forms of P5CS also exhibit diffused distribution. Multimerization (but not the catalytic activity) of P5CS regulates its localization. P5CS mutant cells have a reduced proliferation rate and are sensitive to cellular stresses. Flies lacking P5CS have reduced eclosion rates. Lipid droplets accumulate in the eyes of the newly eclosed P5CS mutant flies, which degenerate with aging. The loss of P5CS in cells leads to abnormal purine metabolism and lipid-droplet accumulation. The reduced lipid-droplet consumption is likely due to decreased expression of the fatty acid transporter, CPT1, and few β-oxidation-related genes following P5CS knockdown. Surprisingly, we found that P5CS is required for mitochondrial respiratory complex organization and that the respiration defects in P5CS knockout cells likely contribute to the metabolic defects in purine synthesis and lipid consumption. This study links amino acid synthesis with mitochondrial respiration and other key metabolic processes, whose imbalance might contribute to P5CS-related disease conditions.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

