A versatile bulk electrotransfection protocol for murine embryonic fibroblasts and iPS cells
- PMID: 32770110
- PMCID: PMC7414887
- DOI: 10.1038/s41598-020-70258-w
A versatile bulk electrotransfection protocol for murine embryonic fibroblasts and iPS cells
Abstract
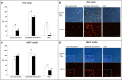
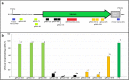
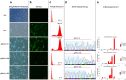
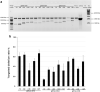
Although electroporation has been widely accepted as the main gene transfer tool, there is still considerable scope to improve the electroporation efficiency of exogenous DNAs into primary cells. Here, we developed a square-wave pulsing protocol using OptiMEM-GlutaMAX for highly efficient transfection of murine embryonic fibroblasts (MEF) and induced pluripotency stem (iPS) cells using reporter genes as well as gRNA/Cas9-encoding plasmids. An electrotransfection efficiency of > 95% was achieved for both MEF and iPS cells using reporter-encoding plasmids. The protocol was efficient for plasmid sizes ranging from 6.2 to 13.5 kb. Inducing the error prone non-homologous end joining repair by gRNA/Cas9 plasmid transfection, a high rate of targeted gene knockouts of up to 98% was produced in transgenic cells carrying a single-copy of Venus reporter. Targeted deletions in the Venus transgene were efficiently (up to 67% deletion rate) performed by co-electroporation of two gRNA-encoding plasmids. We introduced a plasmid electrotransfection protocol which is straight-forward, cost-effective, and efficient for CRISPRing murine primary cells. This protocol is promising to make targeted genetic engineering using the CRISPR/Cas9 plasmid system.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
An Electrochemical Protocol for CRISPR-Mediated Gene-Editing of Sheep Embryonic Fibroblast Cells.Cells Tissues Organs. 2023;212(2):176-184. doi: 10.1159/000521128. Epub 2021 Nov 25. Cells Tissues Organs. 2023. PMID: 34823242
-
Efficient Editing of the Nuclear APT Reporter Gene in Chlamydomonas reinhardtii via Expression of a CRISPR-Cas9 Module.Int J Mol Sci. 2019 Mar 12;20(5):1247. doi: 10.3390/ijms20051247. Int J Mol Sci. 2019. PMID: 30871076 Free PMC article.
-
Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair.Nucleic Acids Res. 2016 May 19;44(9):e85. doi: 10.1093/nar/gkw064. Epub 2016 Feb 4. Nucleic Acids Res. 2016. PMID: 26850641 Free PMC article.
-
CRISPR Knockouts in Ciona Embryos.Adv Exp Med Biol. 2018;1029:141-152. doi: 10.1007/978-981-10-7545-2_13. Adv Exp Med Biol. 2018. PMID: 29542087 Free PMC article. Review.
-
Versatile and precise gene-targeting strategies for functional studies in mammalian cell lines.Methods. 2017 May 15;121-122:45-54. doi: 10.1016/j.ymeth.2017.05.003. Epub 2017 May 10. Methods. 2017. PMID: 28499832 Review.
Cited by
-
Efficient gene editing of BMP15, GDF9, and MSTN-but not the imprinted CLPG gene-in goat embryos via electrotransfection and handmade cloning.Funct Integr Genomics. 2025 Jul 10;25(1):150. doi: 10.1007/s10142-025-01644-8. Funct Integr Genomics. 2025. PMID: 40637794
-
CRISPR/Cas9-mediated targeted knock-in of large constructs using nocodazole and RNase HII.Sci Rep. 2023 Feb 15;13(1):2690. doi: 10.1038/s41598-023-29789-1. Sci Rep. 2023. PMID: 36792645 Free PMC article.
-
Effect of degeneration stage on non-viral tissue transfection of rd10 retina ex vivo.Mol Ther Nucleic Acids. 2025 Jul 1;36(3):102616. doi: 10.1016/j.omtn.2025.102616. eCollection 2025 Sep 9. Mol Ther Nucleic Acids. 2025. PMID: 40704026 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

