A combined risk score enhances prediction of type 1 diabetes among susceptible children
- PMID: 32770166
- PMCID: PMC7556983
- DOI: 10.1038/s41591-020-0930-4
A combined risk score enhances prediction of type 1 diabetes among susceptible children
Erratum in
-
Author Correction: A combined risk score enhances prediction of type 1 diabetes among susceptible children.Nat Med. 2022 Mar;28(3):599. doi: 10.1038/s41591-021-01631-z. Nat Med. 2022. PMID: 35165454 No abstract available.
Abstract
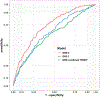
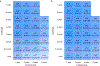
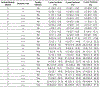
Type 1 diabetes (T1D)-an autoimmune disease that destroys the pancreatic islets, resulting in insulin deficiency-often begins early in life when islet autoantibody appearance signals high risk1. However, clinical diabetes can follow in weeks or only after decades, and is very difficult to predict. Ketoacidosis at onset remains common2,3 and is most severe in the very young4,5, in whom it can be life threatening and difficult to treat6-9. Autoantibody surveillance programs effectively prevent most ketoacidosis10-12 but require frequent evaluations whose expense limits public health adoption13. Prevention therapies applied before onset, when greater islet mass remains, have rarely been feasible14 because individuals at greatest risk of impending T1D are difficult to identify. To remedy this, we sought accurate, cost-effective estimation of future T1D risk by developing a combined risk score incorporating both fixed and variable factors (genetic, clinical and immunological) in 7,798 high-risk children followed closely from birth for 9.3 years. Compared with autoantibodies alone, the combined model dramatically improves T1D prediction at ≥2 years of age over horizons up to 8 years of age (area under the receiver operating characteristic curve ≥ 0.9), doubles the estimated efficiency of population-based newborn screening to prevent ketoacidosis, and enables individualized risk estimates for better prevention trial selection.
Conflict of interest statement
Figures



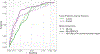




Similar articles
-
A Type 1 Diabetes Genetic Risk Score Predicts Progression of Islet Autoimmunity and Development of Type 1 Diabetes in Individuals at Risk.Diabetes Care. 2018 Sep;41(9):1887-1894. doi: 10.2337/dc18-0087. Epub 2018 Jul 12. Diabetes Care. 2018. PMID: 30002199 Free PMC article.
-
Dynamics of Islet Autoantibodies During Prospective Follow-Up From Birth to Age 15 Years.J Clin Endocrinol Metab. 2020 Dec 1;105(12):e4638-51. doi: 10.1210/clinem/dgaa624. J Clin Endocrinol Metab. 2020. PMID: 32882033 Free PMC article.
-
Extremely Early Appearance of Islet Autoantibodies in Genetically Susceptible Children.Pediatr Diabetes. 2023 Dec 11;2023:9973135. doi: 10.1155/2023/9973135. eCollection 2023. Pediatr Diabetes. 2023. PMID: 40303239 Free PMC article.
-
The Multifactorial Progression from the Islet Autoimmunity to Type 1 Diabetes in Children.Int J Mol Sci. 2021 Jul 13;22(14):7493. doi: 10.3390/ijms22147493. Int J Mol Sci. 2021. PMID: 34299114 Free PMC article. Review.
-
Islet Autoantibodies.Curr Diab Rep. 2016 Jun;16(6):53. doi: 10.1007/s11892-016-0738-2. Curr Diab Rep. 2016. PMID: 27112957 Review.
Cited by
-
Identification of type 1 diabetes risk phenotypes using an outcome-guided clustering analysis.Diabetologia. 2024 Nov;67(11):2507-2517. doi: 10.1007/s00125-024-06246-w. Epub 2024 Aug 6. Diabetologia. 2024. PMID: 39103721 Free PMC article.
-
Clinical and experimental treatment of type 1 diabetes.Clin Exp Immunol. 2022 Dec 15;210(2):105-113. doi: 10.1093/cei/uxac077. Clin Exp Immunol. 2022. PMID: 35980300 Free PMC article. Review.
-
Precision medicine in type 1 diabetes.Diabetologia. 2022 Nov;65(11):1854-1866. doi: 10.1007/s00125-022-05778-3. Epub 2022 Aug 22. Diabetologia. 2022. PMID: 35994083 Free PMC article. Review.
-
Diabetic Ketoacidosis at Manifestation of Type 1 Diabetes in Childhood and Adolescence—Incidence and Risk Factors.Dtsch Arztebl Int. 2021 Jun 4;118(22):367-372. doi: 10.3238/arztebl.m2021.0133. Epub 2021 Jun 4. Dtsch Arztebl Int. 2021. PMID: 34250891 Free PMC article.
-
The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository in the AIRR Data Commons: a practical guide for access, use and contributions through the Type 1 Diabetes AIRR Consortium.Diabetologia. 2025 Jan;68(1):186-202. doi: 10.1007/s00125-024-06298-y. Epub 2024 Oct 29. Diabetologia. 2025. PMID: 39467874 Free PMC article.
References
References Main Text
Methods-Only References
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK063821/DK/NIDDK NIH HHS/United States
- UC4 DK063863/DK/NIDDK NIH HHS/United States
- 17/0005757/DUK_/Diabetes UK/United Kingdom
- HHSN267200700014C/DK/NIDDK NIH HHS/United States
- 16/0005529/DUK_/Diabetes UK/United Kingdom
- U01 DK063790/DK/NIDDK NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 3-SRA-2019-827-S-B/JDRF/Breakthrough T1D/United States
- P30 DK017047/DK/NIDDK NIH HHS/United States
- UL1 TR000064/TR/NCATS NIH HHS/United States
- U01 DK063836/DK/NIDDK NIH HHS/United States
- U01 DK063829/DK/NIDDK NIH HHS/United States
- U01 DK063865/DK/NIDDK NIH HHS/United States
- UC4 DK095300/DK/NIDDK NIH HHS/United States
- UC4 DK063861/DK/NIDDK NIH HHS/United States
- UC4 DK063829/DK/NIDDK NIH HHS/United States
- UC4 DK063821/DK/NIDDK NIH HHS/United States
- UC4 DK117483/DK/NIDDK NIH HHS/United States
- UC4 DK063836/DK/NIDDK NIH HHS/United States
- UC4 DK112243/DK/NIDDK NIH HHS/United States
- U01 DK124166/DK/NIDDK NIH HHS/United States
- U01 DK063861/DK/NIDDK NIH HHS/United States
- UC4 DK063865/DK/NIDDK NIH HHS/United States
- U01 DK063863/DK/NIDDK NIH HHS/United States
- UC4 DK106955/DK/NIDDK NIH HHS/United States
- UC4 DK100238/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

