Joint embedding: A scalable alignment to compare individuals in a connectivity space
- PMID: 32771618
- PMCID: PMC7779372
- DOI: 10.1016/j.neuroimage.2020.117232
Joint embedding: A scalable alignment to compare individuals in a connectivity space
Abstract
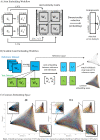
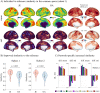
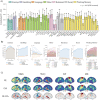
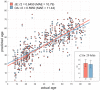
A common coordinate space enabling comparison across individuals is vital to understanding human brain organization and individual differences. By leveraging dimensionality reduction algorithms, high-dimensional fMRI data can be represented in a low-dimensional space to characterize individual features. Such a representative space encodes the functional architecture of individuals and enables the observation of functional changes across time. However, determining comparable functional features across individuals in resting-state fMRI in a way that simultaneously preserves individual-specific connectivity structure can be challenging. In this work we propose scalable joint embedding to simultaneously embed multiple individual brain connectomes within a common space that allows individual representations across datasets to be aligned. Using Human Connectome Project data, we evaluated the joint embedding approach by comparing it to the previously established orthonormal alignment model. Alignment using joint embedding substantially increased the similarity of functional representations across individuals while simultaneously capturing their distinct profiles, allowing individuals to be more discriminable from each other. Additionally, we demonstrated that the common space established using resting-state fMRI provides a better overlap of task-activation across participants. Finally, in a more challenging scenario - alignment across a lifespan cohort aged from 6 to 85 - joint embedding provided a better prediction of age (r2 = 0.65) than the prior alignment model. It facilitated the characterization of functional trajectories across lifespan. Overall, these analyses establish that joint embedding can simultaneously capture individual neural representations in a common connectivity space aligning functional data across participants and populations and preserve individual specificity.
Keywords: Common space; Functional alignment; Functional gradient; Individual differences; Joint embedding; Lifespan.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest None.
Figures


References
-
- Arroyo, J., Athreya, A., Cape, J., Chen, G., Priebe, C.E., Vogelstein, J.T., 2019. Inference for multiple heterogeneous networks with a common invariant subspace. arXiv. https://doi.org/1906.10026. - PMC - PubMed
-
- Baumgartner C.F., Kolbitsch C., McClelland J.R., Rueckert D., King A.P. Groupwise simultaneous manifold alignment for high-resolution dynamic MR imaging of respiratory motion. Inf. Process. Med. Imaging. 2013;23:232–243. - PubMed
-
- Bridgeford, E.W., Wang, S., Yang, Z., Wang, Z., Xu, T., Craddock, C., Kiar, G., Gray-Roncal, W., Priebe, C.E., Caffo, B., Milham, M., Zuo, X.-N., Consortium for Reliability and Reproduciblity, Vogelstein, J.T., 2019. Optimal Experimental Design for Big Data: Applications in Brain Imaging. bioRxiv.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

