Circulating miRNAs as Biomarkers for CYP2B6 Enzyme Activity
- PMID: 32772362
- PMCID: PMC9007394
- DOI: 10.1002/cpt.2018
Circulating miRNAs as Biomarkers for CYP2B6 Enzyme Activity
Abstract
The CYP2B6 gene is highly polymorphic and its activity shows wide interindividual variability. However, substantial variability in CYP2B6 activity remains unexplained by the known CYP2B6 genetic variations. Circulating, cell-free micro RNAs (miRNAs) may serve as biomarkers of hepatic enzyme activity. CYP2B6 activity in 72 healthy volunteers was determined using the disposition of efavirenz as a probe drug. Circulating miRNA expression was quantified from baseline plasma samples. A linear model consisting of the effects of miRNA expression, genotype-determined metabolizer status, and demographic information was developed to predict CYP2B6 activity. Expression of 2,510 miRNAs were quantified out of which 7 miRNAs, together with the CYP2B6-genotypic metabolizer status and demographics, was shown to be predictive markers for CYP2B6 activity. The reproducibility of the model was evaluated by cross-validation. The average Pearson's correlation (R) between the predicted and observed maximum plasma concentration (Cmax ) ratios of efavirenz and its metabolite-8-OH efavirenz using the linear model with all features (7 miRNA + metabolizer status + age + sex + race) was 0.6702. Similar results were also observed using area under the curve (AUC) ratios (Pearson correlation's R = 0.6035). Thus, at least 36% (R2 ) of the variability of in vivo CYP2B6 activity was explained using this model. This is a significant improvement over the models using only the genotype-based metabolizer status or the demographic information, which explained only 6% or less of the variability of in vivo CYP2B6 activity. Our results, therefore, demonstrate that circulating plasma miRNAs can be valuable biomarkers for in vivo CYP2B6 activity.
© 2020 The Authors Clinical Pharmacology & Therapeutics © 2020 American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Conflict of Interest: The authors declared no competing interests for this work.
Figures



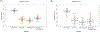
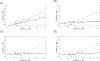
Similar articles
-
CYP2B6 Genetic Polymorphisms, Depression, and Viral Suppression in Adults Living with HIV Initiating Efavirenz-Containing Antiretroviral Therapy Regimens in Uganda: Pooled Analysis of Two Prospective Studies.AIDS Res Hum Retroviruses. 2018 Nov;34(11):982-992. doi: 10.1089/AID.2018.0062. Epub 2018 Aug 15. AIDS Res Hum Retroviruses. 2018. PMID: 29973058 Free PMC article.
-
Meta-analysis of the associations of CYP2B6-516G>T polymorphisms with efavirenz-induced central nervous system side effects and virological outcome in HIV-infected adults.Pharmacogenomics J. 2020 Apr;20(2):246-259. doi: 10.1038/s41397-019-0112-2. Epub 2019 Oct 21. Pharmacogenomics J. 2020. PMID: 31636355
-
Population Pharmacokinetic Modeling To Estimate the Contributions of Genetic and Nongenetic Factors to Efavirenz Disposition.Antimicrob Agents Chemother. 2016 Dec 27;61(1):e01813-16. doi: 10.1128/AAC.01813-16. Print 2017 Jan. Antimicrob Agents Chemother. 2016. PMID: 27799204 Free PMC article. Clinical Trial.
-
Dosage Optimization of Efavirenz Based on a Population Pharmacokinetic-Pharmacogenetic Model of HIV-infected Patients in Thailand.Clin Ther. 2020 Jul;42(7):1234-1245. doi: 10.1016/j.clinthera.2020.04.013. Epub 2020 May 22. Clin Ther. 2020. PMID: 32451120
-
Association of CYP2B6 genetic polymorphisms with bupropion and hydroxybupropion exposure: A systematic review and meta-analysis.Pharmacotherapy. 2022 Jan;42(1):34-44. doi: 10.1002/phar.2644. Epub 2021 Nov 23. Pharmacotherapy. 2022. PMID: 34752647
Cited by
-
Novel Approaches to Characterize Individual Drug Metabolism and Advance Precision Medicine.Drug Metab Dispos. 2023 Oct;51(10):1238-1253. doi: 10.1124/dmd.122.001066. Epub 2023 Jul 7. Drug Metab Dispos. 2023. PMID: 37419681 Free PMC article. Review.
-
Application of Machine Learning in Translational Medicine: Current Status and Future Opportunities.AAPS J. 2021 May 18;23(4):74. doi: 10.1208/s12248-021-00593-x. AAPS J. 2021. PMID: 34008139 Free PMC article.
References
-
- Desta Z et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8, 547–58 (2007). - PubMed
-
- Eap CB et al. Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin Pharmacol Ther 81, 719–28 (2007). - PubMed
-
- Nakajima M et al. Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenetics and Genomics 17, 431–45 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

