Alternative Applications of Genotyping Array Data Using Multivariant Methods
- PMID: 32773169
- PMCID: PMC7572808
- DOI: 10.1016/j.tig.2020.07.006
Alternative Applications of Genotyping Array Data Using Multivariant Methods
Abstract
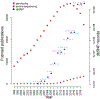
One of the forerunners that pioneered the revolution of high-throughput genomic technologies is the genotyping microarray technology, which can genotype millions of single-nucleotide variants simultaneously. Owing to apparent benefits, such as high speed, low cost, and high throughput, the genotyping array has gained lasting applications in genome-wide association studies (GWAS) and thus accumulated an enormous amount of data. Empowered by continuous manufactural upgrades and analytical innovation, unconventional applications of genotyping array data have emerged to address more diverse genetic problems, holding promise of boosting genetic research into human diseases through the re-mining of the rich accumulated data. Here, we review several unconventional genotyping array analysis techniques that have been built on the idea of large-scale multivariant analysis and provide empirical application examples. These unconventional outcomes of genotyping arrays include polygenic score, runs of homozygosity (ROH)/heterozygosity ratio, distant pedigree computation, and mitochondrial DNA (mtDNA) copy number inference.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

