Comparative Genomics Discloses the Uniqueness and the Biosynthetic Potential of the Marine Cyanobacterium Hyella patelloides
- PMID: 32774329
- PMCID: PMC7381351
- DOI: 10.3389/fmicb.2020.01527
Comparative Genomics Discloses the Uniqueness and the Biosynthetic Potential of the Marine Cyanobacterium Hyella patelloides
Abstract
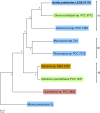
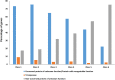
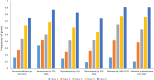
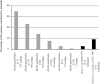
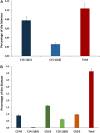
Baeocytous cyanobacteria (Pleurocapsales/Subsection II) can thrive in a wide range of habitats on Earth but, compared to other cyanobacterial lineages, they remain poorly studied at genomic level. In this study, we sequenced the first genome from a member of the Hyella genus - H. patelloides LEGE 07179, a recently described species isolated from the Portuguese foreshore. This genome is the largest of the thirteen baeocyte-forming cyanobacterial genomes sequenced so far, and diverges from the most closely related strains. Comparative analysis revealed strain-specific genes and horizontal gene transfer events between H. patelloides and its closest relatives. Moreover, H. patelloides genome is distinctive by the number and diversity of natural product biosynthetic gene clusters (BGCs). The majority of these clusters are strain-specific BGCs with a high probability of synthesizing novel natural products. One BGC was identified as being putatively involved in the production of terminal olefin. Our results showed that, H. patelloides produces hydrocarbon with C15 chain length, and synthesizes C14, C16, and C18 fatty acids exceeding 4% of the dry cell weight. Overall, our data contributed to increase the information on baeocytous cyanobacteria, and shed light on H. patelloides evolution, phylogeny and natural product biosynthetic potential.
Keywords: Hyella; biosynthetic gene clusters; cyanobacteria; genome; natural products.
Copyright © 2020 Brito, Vieira, Vieira, Zhu, Leão, Ramos, Lu, Vasconcelos, Gugger and Tamagnini.
Figures


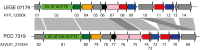
Similar articles
-
Final Destination? Pinpointing Hyella disjuncta sp. nov. PCC 6712 (Cyanobacteria) Based on Taxonomic Aspects, Multicellularity, Nitrogen Fixation and Biosynthetic Gene Clusters.Life (Basel). 2021 Sep 3;11(9):916. doi: 10.3390/life11090916. Life (Basel). 2021. PMID: 34575065 Free PMC article.
-
Isolation and Biosynthesis of Hyellamide, a Glycosylated N-Acyltyrosine Derivative, from the Cyanobacterium Hyella patelloides LEGE 07179.J Nat Prod. 2025 Mar 28;88(3):850-856. doi: 10.1021/acs.jnatprod.4c00968. Epub 2025 Feb 26. J Nat Prod. 2025. PMID: 40009566 Free PMC article.
-
Description of new genera and species of marine cyanobacteria from the Portuguese Atlantic coast.Mol Phylogenet Evol. 2017 Jun;111:18-34. doi: 10.1016/j.ympev.2017.03.006. Epub 2017 Mar 6. Mol Phylogenet Evol. 2017. PMID: 28279808
-
Exploring cyanobacterial genomes for natural product biosynthesis pathways.Mar Genomics. 2015 Jun;21:1-12. doi: 10.1016/j.margen.2014.11.009. Epub 2014 Dec 5. Mar Genomics. 2015. PMID: 25482899 Review.
-
Heterologous production of cyanobacterial compounds.J Ind Microbiol Biotechnol. 2021 Jun 4;48(3-4):kuab003. doi: 10.1093/jimb/kuab003. J Ind Microbiol Biotechnol. 2021. PMID: 33928376 Free PMC article. Review.
Cited by
-
Final Destination? Pinpointing Hyella disjuncta sp. nov. PCC 6712 (Cyanobacteria) Based on Taxonomic Aspects, Multicellularity, Nitrogen Fixation and Biosynthetic Gene Clusters.Life (Basel). 2021 Sep 3;11(9):916. doi: 10.3390/life11090916. Life (Basel). 2021. PMID: 34575065 Free PMC article.
-
Draft genome and description of Waterburya agarophytonicola gen. nov. sp. nov. (Pleurocapsales, Cyanobacteria): a seaweed symbiont.Antonie Van Leeuwenhoek. 2021 Dec;114(12):2189-2203. doi: 10.1007/s10482-021-01672-x. Epub 2021 Oct 21. Antonie Van Leeuwenhoek. 2021. PMID: 34674103 Free PMC article.
-
Isolation and Biosynthesis of Hyellamide, a Glycosylated N-Acyltyrosine Derivative, from the Cyanobacterium Hyella patelloides LEGE 07179.J Nat Prod. 2025 Mar 28;88(3):850-856. doi: 10.1021/acs.jnatprod.4c00968. Epub 2025 Feb 26. J Nat Prod. 2025. PMID: 40009566 Free PMC article.
-
Cyanotoxins and Other Bioactive Compounds from the Pasteur Cultures of Cyanobacteria (PCC).Toxins (Basel). 2023 Jun 9;15(6):388. doi: 10.3390/toxins15060388. Toxins (Basel). 2023. PMID: 37368689 Free PMC article. Review.
References
-
- Al-Thukair A. A. (2011). Calculating boring rate of endolithic cyanobacteria Hyella immanis under laboratory conditions. Int. Biodeter. Biodegr. 65 664–667. 10.1016/j.ibiod.2011.03.009 - DOI
-
- Al−Thukair A. A., Golubic S. (1991). New endolithic cyanobacteria from the arabian gulf. I. Hyella immanis SP. NOV. 1. J. Phycol. 27 766–780. 10.1111/j.0022-3646.1991.00766.x - DOI
LinkOut - more resources
Full Text Sources

