A novel numerical representation for proteins: Three-dimensional Chaos Game Representation and its Extended Natural Vector
- PMID: 32774785
- PMCID: PMC7390779
- DOI: 10.1016/j.csbj.2020.07.004
A novel numerical representation for proteins: Three-dimensional Chaos Game Representation and its Extended Natural Vector
Abstract
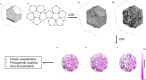
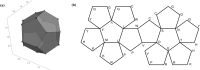
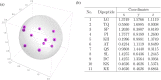
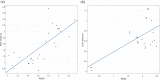
Chaos Game Representation (CGR) was first proposed to be an image representation method of DNA and have been extended to the case of other biological macromolecules. Compared with the CGR images of DNA, where DNA sequences are converted into a series of points in the unit square, the existing CGR images of protein are not so elegant in geometry and the implications of the distribution of points in the CGR image are not so obvious. In this study, by naturally distributing the twenty amino acids on the vertices of a regular dodecahedron, we introduce a novel three-dimensional image representation of protein sequences with CGR method. We also associate each CGR image with a vector in high dimensional Euclidean space, called the extended natural vector (ENV), in order to analyze the information contained in the CGR images. Based on the results of protein classification and phylogenetic analysis, our method could serve as a precise method to discover biological relationships between proteins.
Keywords: Chaos Game Representation; Extended Natural Vector; Protein classification; Three-dimensional CGR.
© 2020 The Author(s).
Figures





References
-
- Rigden D.J. Springer; Netherlands: 2017. From protein structure to function with bioinformatics.
LinkOut - more resources
Full Text Sources
