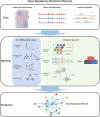
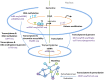
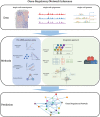
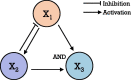
Integration of single-cell multi-omics for gene regulatory network inference
- PMID: 32774787
- PMCID: PMC7385034
- DOI: 10.1016/j.csbj.2020.06.033
Integration of single-cell multi-omics for gene regulatory network inference
Abstract
The advancement of single-cell sequencing technology in recent years has provided an opportunity to reconstruct gene regulatory networks (GRNs) with the data from thousands of single cells in one sample. This uncovers regulatory interactions in cells and speeds up the discoveries of regulatory mechanisms in diseases and biological processes. Therefore, more methods have been proposed to reconstruct GRNs using single-cell sequencing data. In this review, we introduce technologies for sequencing single-cell genome, transcriptome, and epigenome. At the same time, we present an overview of current GRN reconstruction strategies utilizing different single-cell sequencing data. Bioinformatics tools were grouped by their input data type and mathematical principles for reader's convenience, and the fundamental mathematics inherent in each group will be discussed. Furthermore, the adaptabilities and limitations of these different methods will also be summarized and compared, with the hope to facilitate researchers recognizing the most suitable tools for them.
Keywords: Gene regulatory network inference; Single-cell multi-omics integration; Single-cell sequencing.
© 2020 The Authors.
Figures
Similar articles
-
Reconstruction of gene regulatory networks from single cell transcriptomic data.Vavilovskii Zhurnal Genet Selektsii. 2024 Dec;28(8):974-981. doi: 10.18699/vjgb-24-104. Vavilovskii Zhurnal Genet Selektsii. 2024. PMID: 39944798 Free PMC article.
-
Gene regulatory network reconstruction: harnessing the power of single-cell multi-omic data.NPJ Syst Biol Appl. 2023 Oct 19;9(1):51. doi: 10.1038/s41540-023-00312-6. NPJ Syst Biol Appl. 2023. PMID: 37857632 Free PMC article. Review.
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
scGRN: a comprehensive single-cell gene regulatory network platform of human and mouse.Nucleic Acids Res. 2024 Jan 5;52(D1):D293-D303. doi: 10.1093/nar/gkad885. Nucleic Acids Res. 2024. PMID: 37889053 Free PMC article.
-
A review on gene regulatory network reconstruction algorithms based on single cell RNA sequencing.Genes Genomics. 2024 Jan;46(1):1-11. doi: 10.1007/s13258-023-01473-8. Epub 2023 Nov 30. Genes Genomics. 2024. PMID: 38032470 Review.
Cited by
-
PMF-GRN: a variational inference approach to single-cell gene regulatory network inference using probabilistic matrix factorization.Genome Biol. 2024 Apr 8;25(1):88. doi: 10.1186/s13059-024-03226-6. Genome Biol. 2024. PMID: 38589899 Free PMC article.
-
scTIGER: A Deep-Learning Method for Inferring Gene Regulatory Networks from Case versus Control scRNA-seq Datasets.Int J Mol Sci. 2023 Aug 28;24(17):13339. doi: 10.3390/ijms241713339. Int J Mol Sci. 2023. PMID: 37686146 Free PMC article.
-
Resolving trained immunity with systems biology.Eur J Immunol. 2021 Apr;51(4):773-784. doi: 10.1002/eji.202048882. Epub 2021 Mar 10. Eur J Immunol. 2021. PMID: 33570164 Free PMC article. Review.
-
Deep learning-based cell-specific gene regulatory networks inferred from single-cell multiome data.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf138. doi: 10.1093/nar/gkaf138. Nucleic Acids Res. 2025. PMID: 40037709 Free PMC article.
-
A large-scale benchmark for network inference from single-cell perturbation data.Commun Biol. 2025 Mar 11;8(1):412. doi: 10.1038/s42003-025-07764-y. Commun Biol. 2025. PMID: 40069299 Free PMC article.
References
-
- Akaike H. Information theory and an extension of the maximum likelihood principle. Selected Papers Hirotugu Akaike (Springer) 1998:199–213.
-
- Aubin-Frankowski P.-C., Vert J.-P. Gene regulation inference from single-cell RNA-seq data with linear differential equations and velocity inference. BioRxiv. 2018 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous