Longevity and the drift barrier: Bridging the gap between Medawar and Hamilton
- PMID: 32774886
- PMCID: PMC7403686
- DOI: 10.1002/evl3.173
Longevity and the drift barrier: Bridging the gap between Medawar and Hamilton
Abstract
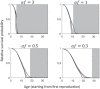
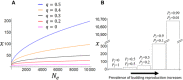
Most organisms have finite life spans. The maximum life span of mammals, for example, is at most some years, decades, or centuries. Why not thousands of years or more? Can we explain and predict maximum life spans theoretically, based on other traits of organisms and associated ecological constraints? Existing theory provides reasons for the prevalence of ageing, but making explicit quantitative predictions of life spans is difficult. Here, I show that there are important unappreciated differences between two backbones of the theory of senescence: Peter Medawar's verbal model, and William Hamilton's subsequent mathematical model. I construct a mathematical model corresponding more closely to Medawar's verbal description, incorporating mutations of large effect and finite population size. In this model, the drift barrier provides a standard by which the limits of natural selection on age-specific mutations can be measured. The resulting model reveals an approximate quantitative explanation for typical maximum life spans. Although maximum life span is expected to increase with population size, it does so extremely slowly, so that even the largest populations imaginable have limited ability to maintain long life spans. Extreme life spans that are observed in some organisms are explicable when indefinite growth or clonal reproduction is included in the model.
Keywords: Evolution of ageing; genetic drift; senescence; theory.
© 2020 The Authors. Evolution Letters published by Wiley Periodicals LLC on behalf of Society for the Study of Evolution (SSE) and European Society for Evolutionary Biology (ESEB).
Conflict of interest statement
The author declares no conflict of interest.
Figures

References
-
- Baudisch, A. 2011. The pace and shape of ageing. Methods Ecol. Evol. 2:375–382.
-
- Caswell, H. 1985. The evolutionary demography of clonal reproduction Pp. 187–224 in Jackson J. B. C., Buss L. W., and Cook R. E., eds. Population biology and evolution of clonal organisms. Yale Univ. Press, New Haven, CT.
-
- Caswell, H. , and Shyu E.. 2017. Senescence, selection gradients and mortality Pp. 56–82 in Shefferson R. P., Jones O. R., and Salguero‐Gómez R., eds. The evolution of senescence in the tree of life. Cambridge Univ. Press, Cambridge, U.K.
LinkOut - more resources
Full Text Sources
Miscellaneous

