Protective anti-prion antibodies in human immunoglobulin repertoires
- PMID: 32776637
- PMCID: PMC7506995
- DOI: 10.15252/emmm.202012739
Protective anti-prion antibodies in human immunoglobulin repertoires
Abstract
Prion immunotherapy may hold great potential, but antibodies against certain PrP epitopes can be neurotoxic. Here, we identified > 6,000 PrP-binding antibodies in a synthetic human Fab phage display library, 49 of which we characterized in detail. Antibodies directed against the flexible tail of PrP conferred neuroprotection against infectious prions. We then mined published repertoires of circulating B cells from healthy humans and found antibodies similar to the protective phage-derived antibodies. When expressed recombinantly, these antibodies exhibited anti-PrP reactivity. Furthermore, we surveyed 48,718 samples from 37,894 hospital patients for the presence of anti-PrP IgGs and found 21 high-titer individuals. The clinical files of these individuals did not reveal any enrichment of specific pathologies, suggesting that anti-PrP autoimmunity is innocuous. The existence of anti-prion antibodies in unbiased human immunological repertoires suggests that they might clear nascent prions early in life. Combined with the reported lack of such antibodies in carriers of disease-associated PRNP mutations, this suggests a link to the low incidence of spontaneous prion diseases in human populations.
Keywords: anti-PrP antibodies; human immunological repertoires; next-generation sequencing; phage display; prion disease.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
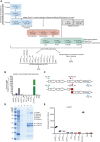
- A
RecPrP23‐231 (blue boxes) was used as target for the first and second round of panning. In the third round, selected phages were panned against recPrP23‐231, recPrP23‐231 under different washing conditions, recPrP fragments (red boxes) and peptides (green boxes) to select for Fabs targeting specific regions of PrP. The DNA preparation from the selected phages was used for NGS of the HCDR3 or ELISA. Negative controls: BSA and Neu (gray boxes).
- B
Bar plot of the NGS counts, relative to recPrP23‐231, for the indicated HCDR3 sequence.
- C
PCR strategy to retrieve the clone of interest. Specific HCDR3_Rw and HCDR3_Fw primers (red arrows) were designed as complementary to the sequence to be rescued. In two separate PCR reactions, the HCDR3_Rw primer was used in combination with the Pal_Fw primer (blue arrow) annealing upstream of the VL, whereas the HCDR3_Fw primer was used in combination with the Flag_RW primer annealing to the His‐tag sequence (blue arrow). The two amplicons were then assembled in a second PCR reaction resulting in the full Fab sequence.
- D
The retrieved Fab clone (FabRTV) was expressed in E. coli and purified by IMAC. The purity of the Fab was analyzed by SDS–PAGE.
- E
FabRTV was tested for its binding specificity to recPrP23‐231 and the indicated PrP fragments and peptides by ELISA. As determined by the NGS analysis (B) and given as an example, FabRTV binds to CC2‐HC92‐120.
- A
Heat maps representing enriched HCDR3 sequences across the different panning sets for the short‐ and long‐HCDR3 (upper and lower panels, respectively) phagemid libraries. HCDR3 sequences were selected based on NGS counts in 100,000 analyzed sequences (Z‐score values; Appendix Table S2) and clustered according to the NGS‐binding profiles. Red and blue: high and low number of NGS counts of the HCDR3 sequence, respectively. Donut charts (the right side) of each heat map indicate the number of clones with NGS‐identified HCDR3 for a predicted PrP epitope.
- B
Bar graphs showing the number of rare (one count in mouse recPrP23‐231 panning) and predominant (count ≧ 20) clones binding to the various PrP regions for the short‐ and long‐HCDR3 libraries.

- A
ELISA titration curves (OD at 450 nm) of Fab3 (blue; left panel) and Fab71 (blue; right panel) against mouse recPrP23‐231 compared to their best affinity‐matured versions, Fab83 and Fab101 (red; EC50 values in Dataset EV1). Curves were fitted by non‐linear least‐squares regression analysis for statistical analysis: log(agonist) vs. response.
- B–E
FT‐peptide competition ELISA to map the epitopes of the indicated Fabs. Peptides that strongly inhibit the binding of the Fabs to recPrP23‐231 are indicated by their residue numbers in the PrP sequence and reflect the respective binding epitopes. Positive control: recPrP23‐231; negative controls: Neu, BSA, and the secondary antibody (2° Ab).

- A, B
Western blot analysis to compare the reactivity of the indicated Fabs to mouse (m) recPrP23‐231 and human (h) recPrP23‐230 (A) and to full‐length and truncated PrPC in BHs of various mouse lines. Actin was used as loading control (B).
- C
ELISA to compare the efficiency of Fab83 and POM1 to detect and quantify wt and mutant PrPC levels in CAD5‐Prnp −/− cells transfected either with wt or mutant PrPC (deletion mutants PrPΔ23‐31 or PrPΔ23‐27; with lysine residues 23, 24, and 27 replaced by alanine; with KKRPK exchanged to KPRKK). All concentrations (% relative to wtPrPC) were determined by interpolating the ELISA signal to a standard curve of mouse recPrP23‐231.

- A
Fab83 coupled beads immunoprecipitated wtPrPC from NBH, but not from BH of PrPΔ25–50 mice lacking the respective epitope. WtPrPC specifically eluted by competition with the Fab83 epitope‐targeting peptide P1 (residues 23–34), but not with the unrelated peptide P15 (top panel). Elution with a SDS buffer was used as nonspecific control (lower panel). Control: IgG‐His. Molecular sizes are presented in kDa.
- B
Same as (A), but for Fab71 coupled beads which immunoprecipitated wtPrPC from NBH, but not from BHs of PrPΔOR mice. WtPrPC specifically eluted with the epitope‐targeting peptide P17 (residues 55–66), but not with the unrelated peptide P2 (top panel).
- C
IgG‐His-Fab100 coupled beads efficiently immunoprecipitated wtPrPC from non‐infectious brain homogenate (NBH) and total wtPrP from brains of prion‐infected mice (prion), but not from brains of Prnp ZH3/ZH3 (ZH3) mice. WtPrP was eluted by competition with the epitope‐targeting peptide P17, but not with the unrelated peptide P2. Eluates were visualized by Western blotting. Control: IgG‐His. Molecular sizes are presented in kDa.
- D
Same as (C) for NBH and prion to confirm the specific immunoprecipitation and detection of PrPSc with the peptide P17 from brains of prion‐infected mice. +PK: digested with PK; −PK: non‐digested with PK; *: molecular weight markers.
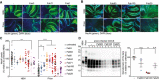
- A
Fluorescence micrographs of tga20 slices chronically exposed to prions and cultured in the presence of either Fab3 (CC123‐50), Fab71 (OR51‐91), or Fab1 (GD) for 45 days. Scale bar: 500 μm.
- B
Same as (A), but in the presence of the affinity‐matured antibodies.
- C
NeuN immunofluorescence coverage of prion‐infected slices cultured in the presence of the Fabs. Antibodies targeting the CC123‐50 and the OR51‐91 afforded protection against neurodegeneration. NBH: non‐infectious brain homogenate. Each dot represents a cerebellar slice for all treatment groups.
Data are indicated as the mean ± sem. Two‐way ANOVA followed by Bonferroni's post hoc test; **P < 0.01; ***P < 0.001; ****P < 0.0001.
- D
Western blot analysis of COCS lysates. Prion‐infected slices treated with Fab100, but not with Fab83 and Fab25 showed reduced levels of PrPSc. n = 3 biological replicates.Data are indicated as mean ± sem. One‐way ANOVA followed by Bonferroni's post hoc test was used for statistical analysis: **P < 0.01, n.s.: not significant.
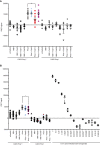
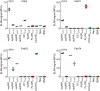
- A
TR‐FRET for the detection of PrPSc in prion‐infected CAD5 Prnp +/+ cells after treatment with selected anti‐PrP Fabs. Treatment of the cells with Fab71 (OR51‐91), but not with Fab3 (CC123‐50) or Fab29 (GD), significantly reduced PrPSc levels compared to untreated, prion‐infected cells.
- B
Treatment of prion‐infected CAD5 Prnp +/+ cells with Fab100 (OR51‐91 binder), but not with Fab83 (CC123‐50 binder), significantly reduced wtPrPSc levels compared to untreated prion‐infected CAD5 Prnp +/+ cells. The dilution range (3 × 10−5% to 1% w/v) of prion‐infected BH is shown on the right site to indicate the linear range of the assay. Negative controls: NBH and CAD5 Prnp −/− cells. Here and in (A), the FRET signal of untreated CAD5 Prnp −/− cells was set to zero. The FRET anti‐PrP antibody pair POM19‐Eu and POM1‐APC was used for detection.

- A
Fab71‐similar HCDR3 sequences in three different NGS datasets (DW: dataset for VH:VL; BB and DK datasets for VH) of human antibody repertoires from healthy donors. Pie chart and stacked bar plot (middle) indicate the occurrence of the identified Fab71 similar HCDR3s: 555 out of 629 identified sequences were only found in one donor, while 74 sequences were shared between donors and occurred in 2 up to six donors. Stacked bar plots (right) report the number of sequences differing to Fab71 HCDR3 by 3, 2 or 1 residues. In total, 13 sequences differed by only one residue. Among them, four sequences were shared between different donors.
- B
Sequence alignment of Fab71 VH3–30 with HCDR3 regions from the different databases that differ from Fab71 by ≤ 3 residues (upper panel). Amino acids differing from Fab71 are highlighted by orange boxes. Lower panel: sequence alignment of Fab71 light‐chain VK3–15 with DK_VL2–14 that is naturally paired with DK_VH3‐07.
- C
Left: Heat map showing the binding specificities (Z‐scores) of selected Fab71 human analogous antibodies to human recPrP23‐230 compared to the negative controls (BSA and Neu). Right: Heat map representing the reactivity (−logEC50) obtained from dose‐dependent ELISA binding curves of the analogous antibodies to human recPrP23‐230. Red: low reactivity; blue: high reactivity.
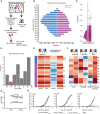
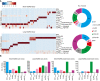
- A
Schematic workflow for the HTS of patient plasma samples for anti‐PrP autoantibodies.
- B
Age pyramid, separated for females and males, of the 37,894 unselected hospital patients tested in the HTS.
- C
Boxplot with half‐violin plot displaying the distribution of autoantibody reactivity in 48,718 plasma samples. Twenty‐seven samples over the reactivity threshold of −log(EC50) ≥ 2 and fitting error < 20% were considered as hits in the primary screen. Black dots correspond to single samples. The boxplot divides the dataset into three quartiles. The box extends from the first to the third quartile values of the dataset, with a line at the median. Whiskers show the range of the data (minimum and maximum).
- D
Frequency distribution of hits based on age.
- E
Heat map representation for the validation of the hits from the primary screen (21 out of 27 hits). Confirmed hits displayed selective reactivity against human recPrP23‐230. Controls: Patients #11–13, ApoE4 and BSA. Red: high reactivity; blue: low reactivity.
- F
Heat maps representing autoantibody clonality of hits analyzed by ELISA. Autoantibodies were mostly constituted by lambda light chains (λ), while the kappa light chains (κ) showed prevalence in the total IgG fraction.
- G
Determination of antibody reactivity (‐log(EC50)) for three individuals at two different time points. Individuals were selected by displaying strong anti‐PrP IgG reactivity in the first test (Time 1). Reactivity was maintained for even more than one year (Time 2).
- H
Purified IgGs from patients with strong anti‐PrP reactivity were tested against PrP variants and BSA (negative control) in dose‐dependent ELISAs. Patient #5 and Patient #6 showed binding mostly to the human recPrP121‐230. Patient #1 displayed a polyreactive pattern with binding to the human recPrP121‐230 and mouse recPrP23–110.
References
-
- Adamson CS, Yao Y, Vasiljevic S, Sy MS, Ren J, Jones IM (2007) Novel single chain antibodies to the prion protein identified by phage display. Virology 358: 166–177 - PubMed
-
- Arndt KM, Muller KM, Pluckthun A (2001) Helix‐stabilized Fv (hsFv) antibody fragments: substituting the constant domains of a Fab fragment for a heterodimeric coiled‐coil domain. J Mol Biol 312: 221–228 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Stiftung Synapsis - Alzheimer Forschung Schweiz AFS/International
- 160672/ERA-Net/International
- Clinical Research Priority Program Small RNAs/International
- 2014/260/SystemsX.ch (Swiss SystemsX.ch)/International
- 2015/320/SystemsX.ch (Swiss SystemsX.ch)/International
- Theodor und Ida Herzog-Egli Foundation/International
- 2017DRI17/Swiss Personalized Health Network (SPHN)/International
- Stavros Niarchos Foundation (SNF)/International
- 670958/EC | H2020 | H2020 Priority Excellent Science | H2020 European Research Council (ERC)/International
- 768415/EC | H2020 | H2020 Priority Excellent Science | H2020 European Research Council (ERC)/International
- 179040/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF)/International
- 183563/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF)/International
- Ono Pharmaceutical (Ono Pharmaceutical Co., Ltd.)/International
- NOMIS Stiftung (NOMIS Foundation)/International
- Donation by Dr. Hans Salvisberg/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

