Holo-Omics: Integrated Host-Microbiota Multi-omics for Basic and Applied Biological Research
- PMID: 32777774
- PMCID: PMC7416341
- DOI: 10.1016/j.isci.2020.101414
Holo-Omics: Integrated Host-Microbiota Multi-omics for Basic and Applied Biological Research
Abstract
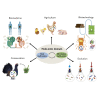
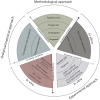
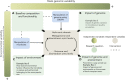
From ontogenesis to homeostasis, the phenotypes of complex organisms are shaped by the bidirectional interactions between the host organisms and their associated microbiota. Current technology can reveal many such interactions by combining multi-omic data from both hosts and microbes. However, exploring the full extent of these interactions requires careful consideration of study design for the efficient generation and optimal integration of data derived from (meta)genomics, (meta)transcriptomics, (meta)proteomics, and (meta)metabolomics. In this perspective, we introduce the holo-omic approach that incorporates multi-omic data from both host and microbiota domains to untangle the interplay between the two. We revisit the recent literature on biomolecular host-microbe interactions and discuss the implementation and current limitations of the holo-omic approach. We anticipate that the application of this approach can contribute to opening new research avenues and discoveries in biomedicine, biotechnology, agricultural and aquacultural sciences, nature conservation, as well as basic ecological and evolutionary research.
Keywords: Evolutionary Biology; Microbiome.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Alberdi A., Aizpurua O., Bohmann K., Zepeda-Mendoza M.L., Gilbert M.T.P. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 2016;31:689–699. - PubMed
-
- Allendorf F.W., Hohenlohe P.A., Luikart G. Genomics and the future of conservation genetics. Nat. Rev. Genet. 2010;11:697–709. - PubMed
-
- Bach J.-F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002;347:911–920. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

