Metagenomics revealing molecular profiling of community structure and metabolic pathways in natural hot springs of the Sikkim Himalaya
- PMID: 32778049
- PMCID: PMC7418396
- DOI: 10.1186/s12866-020-01923-3
Metagenomics revealing molecular profiling of community structure and metabolic pathways in natural hot springs of the Sikkim Himalaya
Abstract
Background: Himalaya is an ecologically pristine environment. The geo-tectonic activities have shaped various environmental niches with diverse microbial populations throughout the Himalayan biosphere region. Albeit, limited information is available in terms of molecular insights into the microbiome, including the uncultured microbes, of the Himalayan habitat. Hence, a vast majority of genomic resources are still under-explored from this region. Metagenome analysis has simplified the extensive in-depth exploration of diverse habitats. In the present study, the culture-independent whole metagenome sequencing methodology was employed for microbial diversity exploration and identification of genes involved in various metabolic pathways in two geothermal springs located at different altitudes in the Sikkim Himalaya.
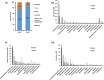
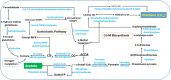
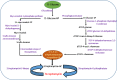
Results: The two hot springs, Polok and Reshi, have distinct abiotic conditions. The average temperature of Polok and Reshi was recorded to be 62 °C and 43 °C, respectively. Both the aquatic habitats have alkaline geochemistry with pH in the range of 7-8. Community profile analysis revealed genomic evidence of plentiful bacteria, with a minute fraction of the archaeal population in hot water reservoirs of Polok and Reshi hot spring. Mesophilic microbes belonging to Proteobacteria and Firmicutes phyla were predominant at both the sites. Polok exhibited an extravagant representation of Chloroflexi, Deinococcus-Thermus, Aquificae, and Thermotogae. Metabolic potential analysis depicted orthologous genes associated with sulfur, nitrogen, and methane metabolism, contributed by the microflora in the hydrothermal system. The genomic information of many novel carbohydrate-transforming enzymes was deciphered in the metagenomic description. Further, the genomic capacity of antimicrobial biomolecules and antibiotic resistance were discerned.
Conclusion: The study provided comprehensive molecular information about the microbial treasury as well as the metabolic features of the two geothermal sites. The thermal aquatic niches were found a potential bioresource of biocatalyst systems for biomass-processing. Overall, this study provides the whole metagenome based insights into the taxonomic and functional profiles of Polok and Reshi hot springs of the Sikkim Himalaya. The study generated a wealth of genomic data that can be explored for the discovery and characterization of novel genes encoding proteins of industrial importance.
Keywords: Antibiotic resistance; CAZymes; Functional potential; Glycosyltransferases; Hot springs; Metagenomics; Taxonomic profiling.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Microbial ecology of two hot springs of Sikkim: Predominate population and geochemistry.Sci Total Environ. 2018 Oct 1;637-638:730-745. doi: 10.1016/j.scitotenv.2018.05.037. Epub 2018 May 11. Sci Total Environ. 2018. PMID: 29758429
-
Baseline metagenome-assembled genome (MAG) data of Sikkim hot springs from Indian Himalayan geothermal belt (IHGB) showcasing its potential CAZymes, and sulfur-nitrogen metabolic activity.World J Microbiol Biotechnol. 2023 May 3;39(7):179. doi: 10.1007/s11274-023-03631-2. World J Microbiol Biotechnol. 2023. PMID: 37133792
-
Comparative metagenomic analyses of a high-altitude Himalayan geothermal spring revealed temperature-constrained habitat-specific microbial community and metabolic dynamics.Arch Microbiol. 2019 Apr;201(3):377-388. doi: 10.1007/s00203-018-01616-6. Epub 2019 Jan 25. Arch Microbiol. 2019. PMID: 30683956
-
Microbiological studies of hot springs in India: a review.Arch Microbiol. 2018 Jan;200(1):1-18. doi: 10.1007/s00203-017-1429-3. Epub 2017 Sep 8. Arch Microbiol. 2018. PMID: 28887679 Review.
-
Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review.Microorganisms. 2021 Jul 9;9(7):1473. doi: 10.3390/microorganisms9071473. Microorganisms. 2021. PMID: 34361908 Free PMC article. Review.
Cited by
-
Prokaryotes of renowned Karlovy Vary (Carlsbad) thermal springs: phylogenetic and cultivation analysis.Environ Microbiome. 2022 Sep 11;17(1):48. doi: 10.1186/s40793-022-00440-2. Environ Microbiome. 2022. PMID: 36089611 Free PMC article.
-
Optimization of thermostable amylolytic enzyme production from Bacillus cereus isolated from a recreational warm spring via Box Behnken design and response surface methodology.Microb Cell Fact. 2025 Apr 19;24(1):87. doi: 10.1186/s12934-025-02709-w. Microb Cell Fact. 2025. PMID: 40253347 Free PMC article.
-
A shotgun approach to explore the bacterial diversity and a brief insight into the glycoside hydrolases of Samiti lake located in the Eastern Himalayas.J Genet Eng Biotechnol. 2022 Dec 5;20(1):162. doi: 10.1186/s43141-022-00444-y. J Genet Eng Biotechnol. 2022. PMID: 36469176 Free PMC article.
-
Thiocapsa, Lutimaribacter, and Delftia Are Major Bacterial Taxa Facilitating the Coupling of Sulfur Oxidation and Nutrient Recycling in the Sulfide-Rich Isinuka Spring in South Africa.Biology (Basel). 2025 May 5;14(5):503. doi: 10.3390/biology14050503. Biology (Basel). 2025. PMID: 40427692 Free PMC article.
-
Metagenomic profiling of antibiotic resistance genes in Red Sea brine pools.Arch Microbiol. 2023 Apr 15;205(5):195. doi: 10.1007/s00203-023-03531-x. Arch Microbiol. 2023. PMID: 37061654
References
-
- Bougnom BP, Zongo C, McNally A, Ricci V, Etoa FX, Thiele Bruhn S, et al. Wastewater used for urban agriculture in West Africa as a reservoir for antibacterial resistance dissemination. J Environ Res. 2019;168:14–24. - PubMed
-
- Kaushal G, Kumar J, Sangwan RS, Singh SP. Metagenomic analysis of geothermal water reservoir sites exploring carbohydrate-related thermozymes. Int J Biol Macromol. 2018;119:882–895. - PubMed
-
- Agarwal N, Narnoliya LK, Singh SP. Characterization of a novel amylosucrase gene from the metagenome of a thermal aquatic habitat, and its use in turanose production from sucrose biomass. Enzym Microb Technol. 2019;131:109372. - PubMed
-
- Joshi N, Sharma M, Singh SP. Characterization of a novel xylanase from an extreme temperature hot spring metagenome for xylooligosaccharide production. Appl Microbiol Biotechnol. 2020;104:4889–901. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

