Distinct and overlapping roles of STAG1 and STAG2 in cohesin localization and gene expression in embryonic stem cells
- PMID: 32778134
- PMCID: PMC7418333
- DOI: 10.1186/s13072-020-00353-9
Distinct and overlapping roles of STAG1 and STAG2 in cohesin localization and gene expression in embryonic stem cells
Abstract
Background: The three-dimensional organization of the genome in the nucleus plays an integral role in many biological processes, including gene expression. The genome is folded into DNA loops that bring together distal regulatory elements and genes. Cohesin, a ring-shaped protein complex, is a major player in the formation of DNA loops. Cohesin is composed of a core trimer and one of two variant STAG subunits, STAG1 or STAG2. It is not understood whether variant STAG proteins give rise to cohesin complexes with distinct functions. Recent studies have begun to characterize the roles of STAG1 and STAG2, with partially contradictory results.
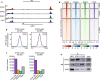
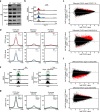
Results: Here, we generate stable single-knockout embryonic stem cell lines to investigate the individual contributions of STAG1 and STAG2 in regulating cohesin chromosomal localization and function. We report both overlapping roles for STAG1 and STAG2 in cohesin localization and somewhat distinct roles in gene expression. STAG1 and STAG2 occupy the same sites across the genome, yet do not exist together in a higher order complex. Despite their shared localization, STAG1 and STAG2 have both distinct and redundant effects on gene expression. Loss of both STAG1 and STAG2 causes widespread transcriptome dysregulation, altered cohesin DNA occupancy, and reduced cell proliferation.
Conclusions: Together, this work reveals the requirement of at least one STAG protein for proper cohesin function. STAG1 and STAG2 have independent roles in cohesin localization and both overlapping and distinct roles in gene expression. The roles of STAG1 and STAG2 in mouse embryonic stem cells may be somewhat different than in other cell types, due to their relative expression levels. These results advance our understanding of the link between mammalian genome organization and gene expression during development and disease contexts.
Keywords: CRISPR/Cas9; CTCF; Cohesin; Enhancers; Gene expression; Promoters; STAG; Stem cell; Transcription.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. - PubMed
-
- Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. - PubMed
-
- Mehta GD, Kumar R, Srivastava S, Ghosh SK. Cohesin: functions beyond sister chromatid cohesion. FEBS Lett. 2013;587(15):2299–2312. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

