Developmental potential of aneuploid human embryos cultured beyond implantation
- PMID: 32778678
- PMCID: PMC7418029
- DOI: 10.1038/s41467-020-17764-7
Developmental potential of aneuploid human embryos cultured beyond implantation
Abstract
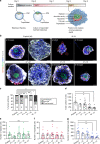
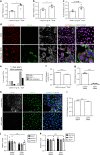
Aneuploidy, the presence of an abnormal number of chromosomes, is a major cause of early pregnancy loss in humans. Yet, the developmental consequences of specific aneuploidies remain unexplored. Here, we determine the extent of post-implantation development of human embryos bearing common aneuploidies using a recently established culture platform. We show that while trisomy 15 and trisomy 21 embryos develop similarly to euploid embryos, monosomy 21 embryos exhibit high rates of developmental arrest, and trisomy 16 embryos display a hypo-proliferation of the trophoblast, the tissue that forms the placenta. Using human trophoblast stem cells, we show that this phenotype can be mechanistically ascribed to increased levels of the cell adhesion protein E-CADHERIN, which lead to premature differentiation and cell cycle arrest. We identify three cases of mosaicism in embryos diagnosed as full aneuploid by pre-implantation genetic testing. Our results present the first detailed analysis of post-implantation development of aneuploid human embryos.
Conflict of interest statement
A.P. is shareholder of IVI-RMA and DIBIMED. E.S. is a consultant for and receives research funding from the Foundation for Embryonic Competence. The remaining authors declare no competing interests.
Figures



References
-
- van den Berg MM, van Maarle MC, van Wely M, Goddijn M. Genetics of early miscarriage. Biochim. Biophys. Acta. 2012;1822:1951–1959. - PubMed
-
- Ljunger E, Cnattingius S, Lundin C, Anneren G. Chromosomal anomalies in first-trimester miscarriages. Acta Obstet. Gynecol. Scand. 2005;84:1103–1107. - PubMed
-
- Strom CM, et al. Analyses of 95 first-trimester spontaneous abortions by chorionic villus sampling and karyotype. J. Assist. Reprod. Genet. 1992;9:458–461. - PubMed
-
- Martinez MC, et al. Cytogenetic analysis of early nonviable pregnancies after assisted reproduction treatment. Fertil. Steril. 2010;93:289–292. - PubMed
-
- Vanneste E, et al. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009;15:577–583. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

