Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents
- PMID: 32778765
- PMCID: PMC7852900
- DOI: 10.1038/s41431-020-00707-7
Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents
Abstract
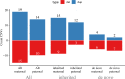
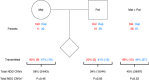
Recurrent copy number variations (CNVs) are common causes of neurodevelopmental disorders (NDDs) and associated with a range of psychiatric traits. These CNVs occur at defined genomic regions that are particularly prone to recurrent deletions and duplications and often exhibit variable expressivity and incomplete penetrance. Robust estimates of the population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders (NDD CNVs) are lacking. Here we perform array-based CNV calling in 12,252 mother-father-child trios from the Norwegian Mother, Father, and Child Cohort Study (MoBa) and analyse the inheritance pattern of 26 recurrent NDD CNVs in 13 genomic regions. We estimate the total prevalence of recurrent NDD CNVs (duplications and deletions) in live-born children to 0.48% (95% C.I.: 0.37-0.62%), i.e., ~1 in 200 newborns has either a deletion or duplication in these NDDs associated regions. Approximately a third of the newborn recurrent NDD CNVs (34%, N = 20/59) are de novo variants. We provide prevalence estimates and inheritance information for each of the 26 NDD CNVs and find higher prevalence than previously reported for 1q21.1 deletions (~1:2000), 15q11.2 duplications (~1:4000), 15q13.3 microdeletions (~1:2500), 16p11.2 proximal microdeletions (~1:2000) and 17q12 deletions (~1:4000) and lower than previously reported prevalence for the 22q11.2 deletion (~1:12,000). In conclusion, our analysis of an unselected and representative population of newborns and their parents provides a clearer picture of the rate of recurrent microdeletions/duplications implicated in neurodevelopmental delay. These results will provide an important resource for genetic diagnostics and counseling.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

