Time-resolved cryo-EM using Spotiton
- PMID: 32778833
- PMCID: PMC7799389
- DOI: 10.1038/s41592-020-0925-6
Time-resolved cryo-EM using Spotiton
Abstract
We present an approach for preparing cryo-electron microscopy (cryo-EM) grids to study short-lived molecular states. Using piezoelectric dispensing, two independent streams of ~50-pl droplets of sample are deposited within 10 ms of each other onto the surface of a nanowire EM grid, and the mixing reaction stops when the grid is vitrified in liquid ethane ~100 ms later. We demonstrate this approach for four biological systems where short-lived states are of high interest.
Figures
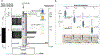
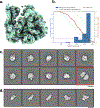
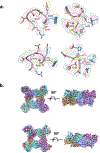



References
-
- Berriman J & Unwin N Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy 56, 241–252 (1994). - PubMed
-
- Unwin N Acetylcholine receptor channel imaged in the open state. Nature 373, 37–43 (1995). - PubMed
-
- White HD, Walker ML & Trinick J A computer-controlled spraying-freezing apparatus for millisecond time-resolution electron cryomicroscopy. J Struct Biol 121, 306–313 (1998). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

