The roles of microRNA in redox metabolism and exercise-mediated adaptation
- PMID: 32780693
- PMCID: PMC7498669
- DOI: 10.1016/j.jshs.2020.03.004
The roles of microRNA in redox metabolism and exercise-mediated adaptation
Abstract
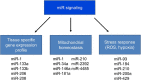
MicroRNAs (miRs) are small regulatory RNA transcripts capable of post-transcriptional silencing of mRNA messages by entering a cellular bimolecular apparatus called RNA-induced silencing complex. miRs are involved in the regulation of cellular processes producing, eliminating or repairing the damage caused by reactive oxygen species, and they are active players in redox homeostasis. Increased mitochondrial biogenesis, function and hypertrophy of skeletal muscle are important adaptive responses to regular exercise. In the present review, we highlight some of the redox-sensitive regulatory roles of miRs.
Keywords: Adaptation; Exercise; MicroRNA; Oxidative damage; Reactive oxygen species; Redox regulation.
Copyright © 2020. Production and hosting by Elsevier B.V.
Figures

References
-
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bhuiyan S.S., Kinoshita S., Wongwarangkana C., Asaduzzaman M., Asakawa S., Watabe S. Evolution of the myosin heavy chain gene MYH14 and its intronic microRNA miR-499: muscle-specific miR-499 expression persists in the absence of the ancestral host gene. BMC Evol Biol. 2013;13:142. doi: 10.1186/1471-2148-13-142. - DOI - PMC - PubMed
-
- Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–299. - PubMed

