Recurrent Rare Copy Number Variants Increase Risk for Esotropia
- PMID: 32780866
- PMCID: PMC7443120
- DOI: 10.1167/iovs.61.10.22
Recurrent Rare Copy Number Variants Increase Risk for Esotropia
Abstract
Purpose: To determine whether rare copy number variants (CNVs) increase risk for comitant esotropia.
Methods: CNVs were identified in 1614 Caucasian individuals with comitant esotropia and 3922 Caucasian controls from Illumina SNP genotyping using two Hidden Markov model (HMM) algorithms, PennCNV and QuantiSNP, which call CNVs based on logR ratio and B allele frequency. Deletions and duplications greater than 10 kb were included. Common CNVs were excluded. Association testing was performed with 1 million permutations in PLINK. Significant CNVs were confirmed with digital droplet polymerase chain reaction (ddPCR). Whole genome sequencing was performed to determine insertion location and breakpoints.
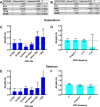
Results: Esotropia patients have similar rates and proportions of CNVs compared with controls but greater total length and average size of both deletions and duplications. Three recurrent rare duplications significantly (P = 1 × 10-6) increase the risk of esotropia: chromosome 2p11.2 (hg19, 2:87428677-87965359), spanning one long noncoding RNA (lncRNA) and two microRNAs (OR 14.16; 95% confidence interval [CI] 5.4-38.1); chromosome 4p15.2 (hg19, 4:25554332-25577184), spanning one lncRNA (OR 11.1; 95% CI 4.6-25.2); chromosome 10q11.22 (hg19, 10:47049547-47703870) spanning seven protein-coding genes, one lncRNA, and four pseudogenes (OR 8.96; 95% CI 5.4-14.9). Overall, 114 cases (7%) and only 28 controls (0.7%) had one of the three rare duplications. No case nor control had more than one of these three duplications.
Conclusions: Rare CNVs are a source of genetic variation that contribute to the genetic risk for comitant esotropia, which is likely polygenic. Future research into the functional consequences of these recurrent duplications may shed light on the pathophysiology of esotropia.
Conflict of interest statement
Disclosure:
Figures






References
-
- Davidson S, Quinn GE. The impact of pediatric vision disorders in adulthood. Pediatrics. 2011; 127: 334–339. - PubMed
-
- Dufier JL, Briard ML, Bonaiti C, Frezal J, Saraux H. Inheritance in the etiology of convergent squint. Ophthalmologica. 1979; 179: 225–234. - PubMed
-
- Podgor MJ, Remaley NA, Chew E. Associations between siblings for esotropia and exotropia. Arch Ophthalmol. 1996; 114: 739–744. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

